Cap analysis of gene expression reveals alternative promoter usage in a rat model of hypertension
- PMID: 34996843
- PMCID: PMC8742872
- DOI: 10.26508/lsa.202101234
Cap analysis of gene expression reveals alternative promoter usage in a rat model of hypertension
Abstract
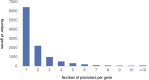
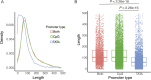
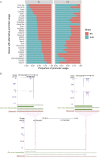
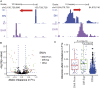
The role of alternative promoter usage in tissue-specific gene expression has been well established; however, its role in complex diseases is poorly understood. We performed cap analysis of gene expression (CAGE) sequencing from the left ventricle of a rat model of hypertension, the spontaneously hypertensive rat (SHR), and a normotensive strain, Brown Norway to understand the role of alternative promoter usage in complex disease. We identified 26,560 CAGE-defined transcription start sites in the rat left ventricle, including 1,970 novel cardiac transcription start sites. We identified 28 genes with alternative promoter usage between SHR and Brown Norway, which could lead to protein isoforms differing at the amino terminus between two strains and 475 promoter switching events altering the length of the 5' UTR. We found that the shift in Insr promoter usage was significantly associated with insulin levels and blood pressure within a panel of HXB/BXH recombinant inbred rat strains, suggesting that hyperinsulinemia due to insulin resistance might lead to hypertension in SHR. Our study provides a preliminary evidence of alternative promoter usage in complex diseases.
© 2022 Dahale et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
MicroRNA-22 and promoter motif polymorphisms at the Chga locus in genetic hypertension: functional and therapeutic implications for gene expression and the pathogenesis of hypertension.Hum Mol Genet. 2013 Sep 15;22(18):3624-40. doi: 10.1093/hmg/ddt213. Epub 2013 May 13. Hum Mol Genet. 2013. PMID: 23674521 Free PMC article.
-
Genetic Variation in Renal Expression of Folate Receptor 1 (Folr1) Gene Predisposes Spontaneously Hypertensive Rats to Metabolic Syndrome.Hypertension. 2016 Feb;67(2):335-41. doi: 10.1161/HYPERTENSIONAHA.115.06158. Epub 2015 Dec 14. Hypertension. 2016. PMID: 26667416
-
An evolutionarily-conserved promoter allele governs HMG-CoA reductase expression in spontaneously hypertensive rat.J Mol Cell Cardiol. 2021 Sep;158:140-152. doi: 10.1016/j.yjmcc.2021.05.017. Epub 2021 May 31. J Mol Cell Cardiol. 2021. PMID: 34081950
-
Use of recombinant inbred strains for evaluation of intermediate phenotypes in spontaneous hypertension.Clin Exp Pharmacol Physiol. 1994 Nov;21(11):903-6. doi: 10.1111/j.1440-1681.1994.tb02463.x. Clin Exp Pharmacol Physiol. 1994. PMID: 7882582 Review.
-
[Cap analysis gene expression: CAGE].Tanpakushitsu Kakusan Koso. 2004 Dec;49(17 Suppl):2688-93. Tanpakushitsu Kakusan Koso. 2004. PMID: 15669240 Review. Japanese. No abstract available.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
