In silico construction of a multiepitope Zika virus vaccine using immunoinformatics tools
- PMID: 34997041
- PMCID: PMC8741764
- DOI: 10.1038/s41598-021-03990-6
In silico construction of a multiepitope Zika virus vaccine using immunoinformatics tools
Abstract
Zika virus (ZIKV) is an arbovirus from the Flaviviridae family and Flavivirus genus. Neurological events have been associated with ZIKV-infected individuals, such as Guillain-Barré syndrome, an autoimmune acute neuropathy that causes nerve demyelination and can induce paralysis. With the increase of ZIKV infection incidence in 2015, malformation and microcephaly cases in newborns have grown considerably, which suggested congenital transmission. Therefore, the development of an effective vaccine against ZIKV became an urgent need. Live attenuated vaccines present some theoretical risks for administration in pregnant women. Thus, we developed an in silico multiepitope vaccine against ZIKV. All structural and non-structural proteins were investigated using immunoinformatics tools designed for the prediction of CD4 + and CD8 + T cell epitopes. We selected 13 CD8 + and 12 CD4 + T cell epitopes considering parameters such as binding affinity to HLA class I and II molecules, promiscuity based on the number of different HLA alleles that bind to the epitopes, and immunogenicity. ZIKV Envelope protein domain III (EDIII) was added to the vaccine construct, creating a hybrid protein domain-multiepitope vaccine. Three high scoring continuous and two discontinuous B cell epitopes were found in EDIII. Aiming to increase the candidate vaccine antigenicity even further, we tested secondary and tertiary structures and physicochemical parameters of the vaccine conjugated to four different protein adjuvants: flagellin, 50S ribosomal protein L7/L12, heparin-binding hemagglutinin, or RS09 synthetic peptide. The addition of the flagellin adjuvant increased the vaccine's predicted antigenicity. In silico predictions revealed that the protein is a probable antigen, non-allergenic and predicted to be stable. The vaccine's average population coverage is estimated to be 87.86%, which indicates it can be administered worldwide. Peripheral Blood Mononuclear Cells (PBMC) of individuals with previous ZIKV infection were tested for cytokine production in response to the pool of CD4 and CD8 ZIKV peptide selected. CD4 + and CD8 + T cells showed significant production of IFN-γ upon stimulation and IL-2 production was also detected by CD8 + T cells, which indicated the potential of our peptides to be recognized by specific T cells and induce immune response. In conclusion, we developed an in silico universal vaccine predicted to induce broad and high-coverage cellular and humoral immune responses against ZIKV, which can be a good candidate for posterior in vivo validation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

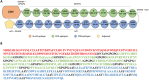


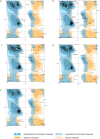
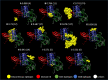

References
-
- Pan American Health Organization. Zika. Zika (2019).
-
- World Health Organization. ZIKA EPIDEMIOLOGY UPDATE July 2019. 1–14 (2019).
-
- Pan American Health Organization. Zika-Epidemiological Report. (2017).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

