Exploring the folding energy landscapes of heme proteins using a hybrid AWSEM-heme model
- PMID: 35000062
- PMCID: PMC8866609
- DOI: 10.1007/s10867-021-09596-3
Exploring the folding energy landscapes of heme proteins using a hybrid AWSEM-heme model
Abstract
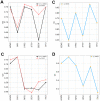
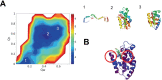
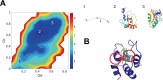
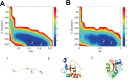
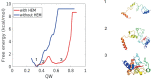
Heme is an active center in many proteins. Here we explore computationally the role of heme in protein folding and protein structure. We model heme proteins using a hybrid model employing the AWSEM Hamiltonian, a coarse-grained forcefield for the protein chain along with AMBER, an all-atom forcefield for the heme. We carefully designed transferable force fields that model the interactions between the protein and the heme. The types of protein-ligand interactions in the hybrid model include thioester covalent bonds, coordinated covalent bonds, hydrogen bonds, and electrostatics. We explore the influence of different types of hemes (heme b and heme c) on folding and structure prediction. Including both types of heme improves the quality of protein structure predictions. The free energy landscape shows that both types of heme can act as nucleation sites for protein folding and stabilize the protein folded state. In binding the heme, coordinated covalent bonds and thioester covalent bonds for heme c drive the heme toward the native pocket. The electrostatics also facilitates the search for the binding site.
Keywords: Forcefield; Heme; Nucleation mechanism; Prediction; Protein folding.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Stokes, G.G. VIII: on the reduction and oxidation of the colouring matter of the blood. Proc. R. Soc. Lond. (13), 355–364 (1864)
-
- Wyman, J.: Allosteric effects in hemoglobin. In: Cold Spring Harbor Symposia on Quantitative Biology, vol. 28, pp. 483–489. Cold Spring Harbor Laboratory Press (1963)
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

