Assessment of tumor burden and response to therapy in patients with colorectal cancer using a quantitative ctDNA test for methylated BCAT1/IKZF1
- PMID: 35000264
- PMCID: PMC9120880
- DOI: 10.1002/1878-0261.13178
Assessment of tumor burden and response to therapy in patients with colorectal cancer using a quantitative ctDNA test for methylated BCAT1/IKZF1
Abstract
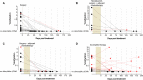
Failure of colorectal cancer (CRC) treatment is due to residual disease, and its timely identification is critical for patient survival. Detecting CRC-associated mutations in patient circulating cell-free DNA is confounded by tumor mutation heterogeneity, requiring primary tumor sequencing to identify relevant mutations. In this study, we assessed BCAT1 and IKZF1 methylation levels to quantify circulating tumor DNA (ctDNA) and investigated whether this method can be used to assess tumor burden and efficacy of therapy. In 175 patients with CRC who were ctDNA-positive pretreatment, ctDNA levels were higher with advancing stage (P < 0.05) and correlated with tumor diameter (r = 0.35, P < 0.001) and volume (r = 0.58, P < 0.01). After completion of treatment (median of 70 days [IQR 49-109] after surgery, +/- radiotherapy, +/- chemotherapy), ctDNA levels were reduced in 98% (47/48) and were undetectable in 88% (42/48) of patients tested. For those with incomplete adjuvant chemotherapy after surgery, roughly half remained ctDNA-positive (11/21, 52.4%). The presence of ctDNA after treatment was associated with disease progression (HR 9.7, 95%CI 2.5-37.6) compared to no ctDNA. Assaying blood for ctDNA methylated in BCAT1/IKZF1 has the potential for identifying residual disease due to treatment failure, informing a potential need for therapy adjustment in advanced disease.
Keywords: BCAT1; IKZF1; circulating tumor DNA; colorectal cancer; efficacy; methylation.
© 2022 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
GPY is a paid consultant of Clinical Genomics. SKP is a paid employee of Clinical Genomics. Other authors have no conflicts of interest to declare.
Figures


References
-
- Niekel MC, Bipat S, Stoker J. Diagnostic imaging of colorectal liver metastases with CT, MR imaging, FDG PET, and/or FDG PET/CT: a meta‐analysis of prospective studies including patients who have not previously undergone treatment. Radiology. 2010;257:674–84. - PubMed
-
- Chao M, Gibbs P. Caution is required before recommending routine carcinoembryonic antigen and imaging follow‐up for patients with early‐stage colon cancer. J Clin Oncol. 2009;27:e279–80. - PubMed
-
- Ruibal Morell A. CEA serum levels in non‐neoplastic disease. Int J Biol Markers. 1992;7:160–6. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

