Forest tree growth is linked to mycorrhizal fungal composition and function across Europe
- PMID: 35001085
- PMCID: PMC9038731
- DOI: 10.1038/s41396-021-01159-7
Forest tree growth is linked to mycorrhizal fungal composition and function across Europe
Abstract
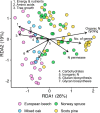
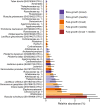
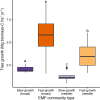
Most trees form symbioses with ectomycorrhizal fungi (EMF) which influence access to growth-limiting soil resources. Mesocosm experiments repeatedly show that EMF species differentially affect plant development, yet whether these effects ripple up to influence the growth of entire forests remains unknown. Here we tested the effects of EMF composition and functional genes relative to variation in well-known drivers of tree growth by combining paired molecular EMF surveys with high-resolution forest inventory data across 15 European countries. We show that EMF composition was linked to a three-fold difference in tree growth rate even when controlling for the primary abiotic drivers of tree growth. Fast tree growth was associated with EMF communities harboring high inorganic but low organic nitrogen acquisition gene proportions and EMF which form contact versus medium-distance fringe exploration types. These findings suggest that EMF composition is a strong bio-indicator of underlying drivers of tree growth and/or that variation of forest EMF communities causes differences in tree growth. While it may be too early to assign causality or directionality, our study is one of the first to link fine-scale variation within a key component of the forest microbiome to ecosystem functioning at a continental scale.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Vittadini C. Monographia lycoperdineorum. Augustae Taurinorum, Torino, 1842.
-
- Frank B. On the nutrition of certain trees by underground fungi based on root symbiosis. Plant Biol. 1885;3:128–45.
-
- Harley J. Problems of mycotrophy. London: Academic Press; 1975.

