Investigation of Cu metal nanoparticles with different morphologies to inhibit SARS-CoV-2 main protease and spike glycoprotein using Molecular Docking and Dynamics Simulation
- PMID: 35001970
- PMCID: PMC8719269
- DOI: 10.1016/j.molstruc.2021.132301
Investigation of Cu metal nanoparticles with different morphologies to inhibit SARS-CoV-2 main protease and spike glycoprotein using Molecular Docking and Dynamics Simulation
Abstract
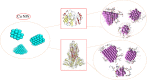
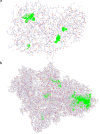
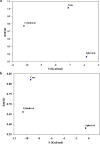
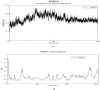
Nowadays, considering the spread of the coronavirus as a global threat, scientific research on this virus through simulation has been increasing. In this study, effect of Cu nanocluster on prevention and control of disease transmission was examined using molecular docking and molecular dynamics simulation studies on the SARS-CoV-2 main protease and spike glycoprotein. The cytotoxicity of different shapes of copper NPs and resonance changes of their surface plasmons on inactivation of the coronavirus was examined in order to control replication of coronavirus through copper NPs, active site of protease and spike glycoprotein. The simulations results showed that interactions of SARS-CoV-2 main protease and spike glycoprotein target and cylindrical and conical copper NPs ligands were more efficient than spherical copper NPs.
Keywords: COVID-19; Copper NPs; Molecular Docking; Molecular Dynamics Simulation; Protease and Spike Glycoprotein Inhibitor.
© 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Figures





















Similar articles
-
Interactions of Co, Cu, and non-metal phthalocyanines with external structures of SARS-CoV-2 using docking and molecular dynamics.Sci Rep. 2022 Feb 28;12(1):3316. doi: 10.1038/s41598-022-07396-w. Sci Rep. 2022. PMID: 35228662 Free PMC article.
-
Computational screening of dual inhibitors from FDA approved antiviral drugs on SARS-CoV-2 spike protein and the main protease using molecular docking approach.Acta Virol. 2021;65(2):160-172. doi: 10.4149/av_2021_208. Acta Virol. 2021. PMID: 34130467
-
In silico prediction of potential inhibitors for the main protease of SARS-CoV-2 using molecular docking and dynamics simulation based drug-repurposing.J Infect Public Health. 2020 Sep;13(9):1210-1223. doi: 10.1016/j.jiph.2020.06.016. Epub 2020 Jun 16. J Infect Public Health. 2020. PMID: 32561274 Free PMC article.
-
Targeting the SARS-CoV-2 spike glycoprotein prefusion conformation: virtual screening and molecular dynamics simulations applied to the identification of potential fusion inhibitors.Virus Res. 2020 Sep;286:198068. doi: 10.1016/j.virusres.2020.198068. Epub 2020 Jun 18. Virus Res. 2020. PMID: 32565126 Free PMC article.
-
Exploring Spike Protein as Potential Target of Novel Coronavirus and to Inhibit the Viability Utilizing Natural Agents.Curr Drug Targets. 2021;22(17):2006-2020. doi: 10.2174/1389450122666210309105820. Curr Drug Targets. 2021. PMID: 33687893 Review.
Cited by
-
In-silico investigation of a novel inhibitors against the antibiotic-resistant Neisseria gonorrhoeae bacteria.Saudi J Biol Sci. 2022 Oct;29(10):103424. doi: 10.1016/j.sjbs.2022.103424. Epub 2022 Aug 22. Saudi J Biol Sci. 2022. PMID: 36091725 Free PMC article.
-
Kinetics of Drug Molecule Interactions with a Newly Developed Nano-Gold-Modified Spike Protein Electrochemical Receptor Sensor.Biosensors (Basel). 2022 Oct 17;12(10):888. doi: 10.3390/bios12100888. Biosensors (Basel). 2022. PMID: 36291025 Free PMC article.
-
Temozolomide Efficacy and Metabolism: The Implicit Relevance of Nanoscale Delivery Systems.Molecules. 2022 May 30;27(11):3507. doi: 10.3390/molecules27113507. Molecules. 2022. PMID: 35684445 Free PMC article. Review.
-
Antibacterial and Antiviral Effects of Ag, Cu and Zn Metals, Respective Nanoparticles and Filter Materials Thereof against Coronavirus SARS-CoV-2 and Influenza A Virus.Pharmaceutics. 2022 Nov 22;14(12):2549. doi: 10.3390/pharmaceutics14122549. Pharmaceutics. 2022. PMID: 36559043 Free PMC article.
-
Can iron, zinc, copper and selenium status be a prognostic determinant in COVID-19 patients?Environ Toxicol Pharmacol. 2022 Oct;95:103937. doi: 10.1016/j.etap.2022.103937. Epub 2022 Jul 23. Environ Toxicol Pharmacol. 2022. PMID: 35882309 Free PMC article. Review.
References
-
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020:579. doi: 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

