Evaluation of PET Degradation Using Artificial Microbial Consortia
- PMID: 35003008
- PMCID: PMC8733400
- DOI: 10.3389/fmicb.2021.778828
Evaluation of PET Degradation Using Artificial Microbial Consortia
Abstract
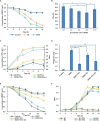
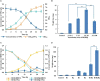
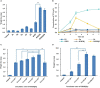
Polyethylene terephthalate (PET) biodegradation is regarded as an environmentally friendly degradation method. In this study, an artificial microbial consortium composed of Rhodococcus jostii, Pseudomonas putida and two metabolically engineered Bacillus subtilis was constructed to degrade PET. First, a two-species microbial consortium was constructed with two engineered B. subtilis that could secrete PET hydrolase (PETase) and monohydroxyethyl terephthalate hydrolase (MHETase), respectively; it could degrade 13.6% (weight loss) of the PET film within 7 days. A three-species microbial consortium was further obtained by adding R. jostii to reduce the inhibition caused by terephthalic acid (TPA), a breakdown product of PET. The weight of PET film was reduced by 31.2% within 3 days, achieving about 17.6% improvement compared with the two-species microbial consortium. Finally, P. putida was introduced to reduce the inhibition caused by ethylene glycol (EG), another breakdown product of PET, obtaining a four-species microbial consortium. With the four-species consortium, the weight loss of PET film reached 23.2% under ambient temperature. This study constructed and evaluated the artificial microbial consortia in PET degradation, which demonstrated the great potential of artificial microbial consortia in the utilization of complex substrates, providing new insights for biodegradation of complex polymers.
Keywords: Polyethylene terephthalate; artificial microbial consortia; biodegradation; ethylene glycol; terephthalic acid.
Copyright © 2021 Qi, Ma, Chang, Li, Ding and Yuan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Current Advances in the Biodegradation and Bioconversion of Polyethylene Terephthalate.Microorganisms. 2021 Dec 26;10(1):39. doi: 10.3390/microorganisms10010039. Microorganisms. 2021. PMID: 35056486 Free PMC article. Review.
-
Development of a Targeted Gene Disruption System in the Poly(Ethylene Terephthalate)-Degrading Bacterium Ideonella sakaiensis and Its Applications to PETase and MHETase Genes.Appl Environ Microbiol. 2021 Aug 26;87(18):e0002021. doi: 10.1128/AEM.00020-21. Epub 2021 Aug 26. Appl Environ Microbiol. 2021. PMID: 34260304 Free PMC article.
-
Environmental Consortium Containing Pseudomonas and Bacillus Species Synergistically Degrades Polyethylene Terephthalate Plastic.mSphere. 2020 Dec 23;5(6):e01151-20. doi: 10.1128/mSphere.01151-20. mSphere. 2020. PMID: 33361127 Free PMC article.
-
[Advances in the structure and function of MHETase].Sheng Wu Gong Cheng Xue Bao. 2024 Sep 25;40(9):2812-2830. doi: 10.13345/j.cjb.230791. Sheng Wu Gong Cheng Xue Bao. 2024. PMID: 39319709 Review. Chinese.
-
Ideonella sakaiensis, PETase, and MHETase: From identification of microbial PET degradation to enzyme characterization.Methods Enzymol. 2021;648:187-205. doi: 10.1016/bs.mie.2020.12.007. Epub 2021 Jan 9. Methods Enzymol. 2021. PMID: 33579403
Cited by
-
Degradation of polyethylene terephthalate (PET) plastics by wastewater bacteria engineered via conjugation.Microb Biotechnol. 2024 Sep;17(9):e70015. doi: 10.1111/1751-7915.70015. Microb Biotechnol. 2024. PMID: 39315602 Free PMC article.
-
Recent Advances in the Chemobiological Upcycling of Polyethylene Terephthalate (PET) into Value-Added Chemicals.J Microbiol Biotechnol. 2023 Jan 28;33(1):1-14. doi: 10.4014/jmb.2208.08048. Epub 2022 Oct 13. J Microbiol Biotechnol. 2023. PMID: 36451300 Free PMC article. Review.
-
Coronas of micro/nano plastics: a key determinant in their risk assessments.Part Fibre Toxicol. 2022 Aug 6;19(1):55. doi: 10.1186/s12989-022-00492-9. Part Fibre Toxicol. 2022. PMID: 35933442 Free PMC article. Review.
-
Adaptive Laboratory Evolution of a Microbial Consortium Enhancing Non-Protein Nitrogen Assimilation for Feed Protein Production.Microorganisms. 2025 Jun 18;13(6):1416. doi: 10.3390/microorganisms13061416. Microorganisms. 2025. PMID: 40572304 Free PMC article.
-
Construction of microbial consortia for microbial degradation of complex compounds.Front Bioeng Biotechnol. 2022 Dec 6;10:1051233. doi: 10.3389/fbioe.2022.1051233. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36561050 Free PMC article. Review.
References
-
- Barth M., Oeser T., Wei R., Then J., Schmidt J., Zimmermann W. (2015). Effect of hydrolysis products on the enzymatic degradation of polyethylene terephthalate nanoparticles by a polyester hydrolase from Thermobifida fusca. Biochem. Eng. J. 93 222–228. 10.1016/j.bej.2014.10.012 - DOI
LinkOut - more resources
Full Text Sources

