Bacteriome of Moist Smokeless Tobacco Products Consumed in India With Emphasis on the Predictive Functional Potential
- PMID: 35003015
- PMCID: PMC8740325
- DOI: 10.3389/fmicb.2021.784841
Bacteriome of Moist Smokeless Tobacco Products Consumed in India With Emphasis on the Predictive Functional Potential
Abstract
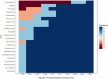
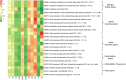
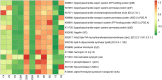
Smokeless tobacco products (STPs) carry assorted microbial population that contributes to carcinogens synthesis like tobacco-specific nitrosamines (TSNAs). Extensive exploration of microbiota-harboring STPs is required to understand their full carcinogenic potential. Here, we applied 16S rRNA gene sequencing to investigate bacteriome present in moist STPs immensely consumed in India (Khaini, Moist-snuff, Qiwam, and Snus). Further, the functional metagenome was speculated by PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) to assign the abundance of genes related to nitrogen metabolism, bacterial toxins, antibiotic drug resistance and other pro-inflammatory molecules. Highly diverse bacterial communities were observed in all moist STPs. Taxonomic analysis revealed a total of 549 genera belonging to four major phyla Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria. Overall, the core bacterial genera Acinetobacter, Bacillus, Prevotella, Acetobacter, Lactobacillus, Paracoccus, Flavobacterium, and Bacteroides were significantly abundant in moist STPs. Elevated moisture-holding products like Moist-snuff and Qiwam harbor rich bacterial species diversity and showed similar bacteriome composition. Furthermore, Qiwam products showed the highest level of genes associated with nitrogen metabolism, antibiotic resistance, toxins, and pro-inflammation (predicted by PICRUSt) which can contribute to the synthesis of TSNAs and induction of oral cancer. The present broad investigation of moist STPs-associated bacteriome prevalence and their detailed metabolic potential will provide novel insight into the oral carcinogenesis induced by STPs.
Keywords: antibiotic-resistance genes; nitrogen metabolism genes; oral cancer; smokeless tobacco products (STPs); smokeless tobacco-associated bacteriome; tobacco-specific nitrosamines (TSNAs); toxins.
Copyright © 2021 Sajid, Srivastava, Kumar, Kumar, Singh and Bharadwaj.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
LinkOut - more resources
Full Text Sources

