Prevalence, Risk Factors, and Genetic Characterization of Extended-Spectrum Beta-Lactamase Escherichia coli Isolated From Healthy Pregnant Women in Madagascar
- PMID: 35003019
- PMCID: PMC8740230
- DOI: 10.3389/fmicb.2021.786146
Prevalence, Risk Factors, and Genetic Characterization of Extended-Spectrum Beta-Lactamase Escherichia coli Isolated From Healthy Pregnant Women in Madagascar
Abstract
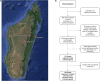
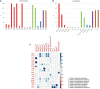
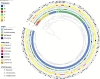
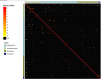
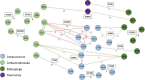
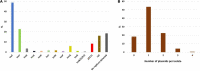
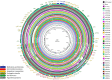
Antimicrobial resistance is a major public health concern worldwide affecting humans, animals and the environment. However, data is lacking especially in developing countries. Thus, the World Health Organization developed a One-Health surveillance project called Tricycle focusing on the prevalence of ESBL-producing Escherichia coli in humans, animals, and the environment. Here we present the first results of the human community component of Tricycle in Madagascar. From July 2018 to April 2019, rectal swabs from 492 pregnant women from Antananarivo, Mahajanga, Ambatondrazaka, and Toamasina were tested for ESBL-E. coli carriage. Demographic, sociological and environmental risk factors were investigated, and E. coli isolates were characterized (antibiotic susceptibility, resistance and virulence genes, plasmids, and genomic diversity). ESBL-E. coli prevalence carriage in pregnant women was 34% varying from 12% (Toamasina) to 65% (Ambatondrazaka). The main risk factor associated with ESBL-E. coli carriage was the rainy season (OR = 2.9, 95% CI 1.3-5.6, p = 0.009). Whole genome sequencing was performed on 168 isolates from 144 participants. bla CTX-M-15 was the most frequent ESBL gene (86%). One isolate was resistant to carbapenems and carried the bla NDM-5 gene. Most isolates belonged to commensalism associated phylogenetic groups A, B1, and C (90%) and marginally to extra-intestinal virulence associated phylogenetic groups B2, D and F (10%). Multi locus sequence typing showed 67 different sequence types gathered in 17 clonal complexes (STc), the most frequent being STc10/phylogroup A (35%), followed distantly by the emerging STc155/phylogroup B1 (7%), STc38/phylogroup D (4%) and STc131/phylogroup B2 (3%). While a wide diversity of clones has been observed, SNP analysis revealed several genetically close isolates (n = 34/168) which suggests human-to-human transmissions. IncY plasmids were found with an unusual prevalence (23%), all carrying a bla CTX-M-15. Most of them (85%) showed substantial homology (≥85%) suggesting a dissemination of IncY ESBL plasmids in Madagascar. This large-scale study reveals a high prevalence of ESBL-E. coli among pregnant women in four cities in Madagascar associated with warmth and rainfall. It shows the great diversity of E. coli disseminating throughout the country but also transmission of specific clones and spread of plasmids. This highlights the urgent need of public-health interventions to control antibiotic resistance in the country.
Keywords: E. coli; Madagascar; digestive carriage; epidemiology; extended-spectrum beta-lactamase (ESBL); whole genome sequencing.
Copyright © 2021 Milenkov, Rasoanandrasana, Rahajamanana, Rakotomalala, Razafindrakoto, Rafalimanana, Ravelomandranto, Ravaoarisaina, Westeel, Petitjean, Mullaert, Clermont, Raskine, Samison, Endtz, Andremont, Denamur, Komurian-Pradel and Armand-Lefevre.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

