Comprehensive Analysis of Pertinent Genes and Pathways in Atrial Fibrillation
- PMID: 35003319
- PMCID: PMC8741379
- DOI: 10.1155/2021/4530180
Comprehensive Analysis of Pertinent Genes and Pathways in Atrial Fibrillation
Abstract
Purpose: Atrial fibrillation (AF) is the most frequent arrhythmia in clinical practice. The pathogenesis of AF is not yet clear. Therefore, exploring the molecular information of AF displays much importance for AF therapy.
Methods: The GSE2240 data were acquired from the Gene Expression Omnibus (GEO) database. The R limma software package was used to screen DEGs. Based on the Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Gene Set Enrichment Analysis (GSEA) databases, we conducted the functions and pathway enrichment analyses. Then, the STRING and Cytoscape software were employed to build Protein-Protein Interaction (PPI) network and screen for hub genes. Finally, we used the Cell Counting Kit-8 (CCK-8) experiment to explore the effect of hub gene knockdown on the proliferation of AF cells.
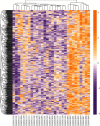
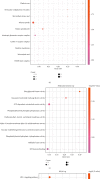
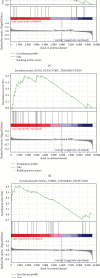
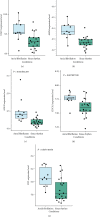
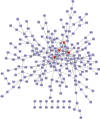
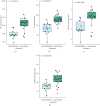
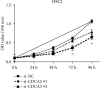
Result: 906 differentially expressed genes (DEGs), including 542 significantly upregulated genes and 364 significantly downregulated genes, were screened in AF. The genes of AF were mainly enriched in vascular endothelial growth factor-activated receptor activity, alanine, regulation of histone deacetylase activity, and HCM. The PPI network constructed of significantly upregulated DEGs contained 404 nodes and 514 edges. Five hub genes, ASPM, DTL, STAT3, ANLN, and CDCA5, were identified through the PPI network. The PPI network constructed by significantly downregulated genes contained 327 nodes and 301 edges. Four hub genes, CDC42, CREB1, AR, and SP1, were identified through this PPI network. The results of CCK-8 experiments proved that knocking down the expression of CDCA5 gene could inhibit the proliferation of H9C2 cells.
Conclusion: Bioinformatics analyses revealed the hub genes and key pathways of AF. These genes and pathways provide information for studying the pathogenesis, treatment, and prognosis of AF and have the potential to become biomarkers in AF treatment.
Copyright © 2021 Yanzhe Wang et al.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures


Similar articles
-
Screening Hub Genes as Prognostic Biomarkers of Hepatocellular Carcinoma by Bioinformatics Analysis.Cell Transplant. 2019 Dec;28(1_suppl):76S-86S. doi: 10.1177/0963689719893950. Epub 2019 Dec 11. Cell Transplant. 2019. PMID: 31822116 Free PMC article.
-
Identification of potential crucial genes in atrial fibrillation: a bioinformatic analysis.BMC Med Genomics. 2020 Jul 18;13(1):104. doi: 10.1186/s12920-020-00754-5. BMC Med Genomics. 2020. PMID: 32682418 Free PMC article.
-
Bioinformatics Analysis Identifies Hub Genes and Molecular Pathways Involved in Sepsis-Induced Myopathy.Med Sci Monit. 2020 Feb 2;26:e919665. doi: 10.12659/MSM.919665. Med Sci Monit. 2020. PMID: 32008037 Free PMC article.
-
Integrated miRNA-mRNA network revealing the key molecular characteristics of ossification of the posterior longitudinal ligament.Medicine (Baltimore). 2020 May 22;99(21):e20268. doi: 10.1097/MD.0000000000020268. Medicine (Baltimore). 2020. PMID: 32481304 Free PMC article.
-
Identification of hub genes in placental dysfunction and recurrent pregnancy loss through transcriptome data mining: A meta-analysis.Taiwan J Obstet Gynecol. 2024 May;63(3):297-306. doi: 10.1016/j.tjog.2024.01.035. Taiwan J Obstet Gynecol. 2024. PMID: 38802191 Review.
Cited by
-
Mathematical Modeling and Computational Prediction of High-Risk Types of Human Papillomaviruses.Comput Math Methods Med. 2022 Jul 21;2022:1515810. doi: 10.1155/2022/1515810. eCollection 2022. Comput Math Methods Med. 2022. PMID: 35912141 Free PMC article.
-
Identification of senescence-related genes for potential therapeutic biomarkers of atrial fibrillation by bioinformatics, human histological validation, and molecular docking.Heliyon. 2024 Sep 4;10(19):e37366. doi: 10.1016/j.heliyon.2024.e37366. eCollection 2024 Oct 15. Heliyon. 2024. PMID: 39381104 Free PMC article.
-
Establishment of Risk Model and Analysis of Immunoinfiltration Based on Mitophagy-Related Associated Genes in Atrial Fibrillation.J Inflamm Res. 2023 Jun 16;16:2561-2583. doi: 10.2147/JIR.S415410. eCollection 2023. J Inflamm Res. 2023. PMID: 37346800 Free PMC article.
-
Decreased METTL3 in atrial myocytes promotes atrial fibrillation.Europace. 2025 Feb 5;27(2):euaf021. doi: 10.1093/europace/euaf021. Europace. 2025. PMID: 39991872 Free PMC article.
References
-
- Moss H. M. Knowledge of Coumadin Use and Vitamin K Interaction in Atrial Fibrillation Patients . Utah State University Utah; 2009.
-
- Friesen A. D. Treatment of atrial fibrillation. Heart . 2007;94
-
- Asadpour P. M., Pardal A. H., Afshar M., Beyranvand M. R. Comparing the serum level of apelin in patients with lone atrial fibrillation and their control group. Pajoohandeh Journal . 2010;15(2):83–87.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

