Updated role of ABA in seed maturation, dormancy, and germination
- PMID: 35003801
- PMCID: PMC8721241
- DOI: 10.1016/j.jare.2021.03.011
Updated role of ABA in seed maturation, dormancy, and germination
Abstract
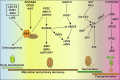
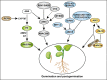
Background: Seed is vital for plant survival and dispersion, however, its development and germination are influenced by various internal and external factors. Abscisic acid (ABA) is one of the most important phytohormones that influence seed development and germination. Until now, impressive progresses in ABA metabolism and signaling pathways during seed development and germination have been achieved. At the molecular level, ABA biosynthesis, degradation, and signaling genes were identified to play important roles in seed development and germination. Additionally, the crosstalk between ABA and other hormones such as gibberellins (GA), ethylene (ET), Brassinolide (BR), and auxin also play critical roles. Although these studies explored some actions and mechanisms by which ABA-related factors regulate seed morphogenesis, dormancy, and germination, the complete network of ABA in seed traits is still unclear.
Aim of review: Presently, seed faces challenges in survival and viability. Due to the vital positive roles in dormancy induction and maintenance, as well as a vibrant negative role in the seed germination of ABA, there is a need to understand the mechanisms of various ABA regulators that are involved in seed dormancy and germination with the updated knowledge and draw a better network for the underlying mechanisms of the ABA, which would advance the understanding and artificial modification of the seed vigor and longevity regulation.
Key scientific concept of review: Here, we review functions and mechanisms of ABA in different seed development stages and seed germination, discuss the current progresses especially on the crosstalk between ABA and other hormones and signaling molecules, address novel points and key challenges (e.g., exploring more regulators, more cofactors involved in the crosstalk between ABA and other phytohormones, and visualization of active ABA in the plant), and outline future perspectives for ABA regulating seed associated traits.
Keywords: Desiccation tolerance; Embryogenesis; Phytohormones; Seed development, Seedling establishment.
© 2021 The Authors. Published by Elsevier B.V. on behalf of Cairo University.
Conflict of interest statement
All the authors in the manuscript have no conflicts of interest.
Figures



References
-
- Graeber K., Nakabayashi K., Miatton E., Leubner-Metzger G., Soppe W.J. Molecular mechanisms of seed dormancy. Plant, Cell Environ. 2012;35(10):1769–1786. - PubMed
-
- Faiza A., Qanmber G., Yonghui L., Shuya M., Lili L., Zuoren Y., et al. Genome-wide identification of Gossypium INDETERMINATE DOMAIN genes and their expression profiles in ovule development and abiotic stress responses. J Cotton Res. 2019
-
- Koornneef M., Bentsink L., Hilhorst H. Seed dormancy and germination. Curr Opin Plant Biol. 2002;5(1):33–36. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

