Advanced high-throughput plant phenotyping techniques for genome-wide association studies: A review
- PMID: 35003802
- PMCID: PMC8721248
- DOI: 10.1016/j.jare.2021.05.002
Advanced high-throughput plant phenotyping techniques for genome-wide association studies: A review
Abstract
Linking phenotypes and genotypes to identify genetic architectures that regulate important traits is crucial for plant breeding and the development of plant genomics. In recent years, genome-wide association studies (GWASs) have been applied extensively to interpret relationships between genes and traits. Successful GWAS application requires comprehensive genomic and phenotypic data from large populations. Although multiple high-throughput DNA sequencing approaches are available for the generation of genomics data, the capacity to generate high-quality phenotypic data is lagging far behind. Traditional methods for plant phenotyping mostly rely on manual measurements, which are laborious, inaccurate, and time-consuming, greatly impairing the acquisition of phenotypic data from large populations. In contrast, high-throughput phenotyping has unique advantages, facilitating rapid, non-destructive, and high-throughput detection, and, in turn, addressing the shortcomings of traditional methods. Aim of Review: This review summarizes the current status with regard to the integration of high-throughput phenotyping and GWAS in plants, in addition to discussing the inherent challenges and future prospects. Key Scientific Concepts of Review: High-throughput phenotyping, which facilitates non-contact and dynamic measurements, has the potential to offer high-quality trait data for GWAS and, in turn, to enhance the unraveling of genetic structures of complex plant traits. In conclusion, high-throughput phenotyping integration with GWAS could facilitate the revealing of coding information in plant genomes.
Keywords: Gene; Imaging; Phenotype; Spectroscopy; Traits.
© 2021 The Authors. Published by Elsevier B.V. on behalf of Cairo University.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
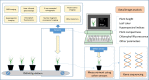
Figures
References
-
- Rebolledo M.C., Pena A.L., Duitama J., Cruz D.F., Dingkuhn M., Grenier C., et al. Combining image analysis, genome wide association studies and different field trials to reveal stable genetic regions related to panicle architecture and the number of spikelets per panicle in rice. Front Plant Sci. 2016;7:1384. doi: 10.3389/fpls.2016.01384. - DOI - PMC - PubMed
-
- Li W.T., Liu C., Liu Y.X., Pu Z.E., Dai S.F., Wang J.R., et al. Meta-analysis of QTL associated with tolerance to abiotic stresses in barley. Euphytica. 2013;189:31–49. doi: 10.1007/s10681-012-0683-3. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

