T and NK cell abundance defines two distinct subgroups of renal cell carcinoma
- PMID: 35003893
- PMCID: PMC8741293
- DOI: 10.1080/2162402X.2021.1993042
T and NK cell abundance defines two distinct subgroups of renal cell carcinoma
Abstract
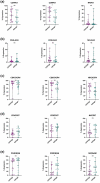
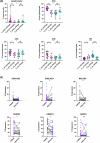
Renal cell carcinoma (RCC) is considered as an immunogenic cancer. Because not all patients respond to current immunotherapies, we aimed to investigate the immunological heterogeneity of RCC tumors. We analyzedthe immunophenotype of the circulating, tumor, and matching adjacent healthy kidney immune cells from 52 nephrectomy patients with multi-parameter flow cytometry. Additionally, we studied the transcriptomic and mutation profiles of 20 clear cell RCC (ccRCC) tumors with bulk RNA sequencing and a customized pan-cancer gene panel. The tumor samples clustered into two distinct subgroups defined by the abundance of intratumoral CD3+ T cells (CD3high, 25/52) and NK cells (NKhigh, 27/52). CD3high tumors had an overall higher frequency of tumor infiltrating lymphocytes and PD-1 expression on the CD8+ T cells compared to NKhigh tumors. The tumor infiltrating T and NK cells had significantly elevated expression levels of LAG-3, PD-1, and HLA-DR compared to the circulating immune cells. Transcriptomic analysis revealed increased immune signaling (IFN-γ, TNF-α via NF-κB, and T cell receptor signaling) and kidney metabolism pathways in the CD3high subgroup. Genomic analysis confirmed the typical ccRCC mutation profile including VHL, PBRM1, and SETD2 mutations, and revealed PBRM1 as a uniquely mutated gene in the CD3high subgroup. Approximately half of the RCC tumors have a high infiltration of NK cells associated with a lower number of tumor infiltrating lymphocytes, lower PD-1 expression, a distinct transcriptomic and mutation profile, providing insights to the immunological heterogeneity of RCC which may impact treatment responses to immunological therapies.
Keywords: NK cell; RCC; T cell; solid tumors; tumor immunology.
© 2022 The Author(s). Published with license by Taylor & Francis Group, LLC.
Conflict of interest statement
SM has received honoraria and research funding from Bristol Myers Squibb, Novartis, and Pfizer outside the submitted work. PJ has received funding from Elypta Ab. AK is currently employed by Novartis. OB has received honoraria from Novartis and Sanofi outside the submitted work. The author(s) declare that there are no other conflicts of interest.
Figures







Similar articles
-
Mutated Von Hippel-Lindau-renal cell carcinoma (RCC) promotes patients specific natural killer (NK) cytotoxicity.J Exp Clin Cancer Res. 2018 Dec 4;37(1):297. doi: 10.1186/s13046-018-0952-7. J Exp Clin Cancer Res. 2018. PMID: 30514329 Free PMC article.
-
Phenotype, cytokine production and cytolytic capacity of fresh (uncultured) tumour-infiltrating T lymphocytes in human renal cell carcinoma.Clin Exp Immunol. 1997 Sep;109(3):501-9. doi: 10.1046/j.1365-2249.1997.4771375.x. Clin Exp Immunol. 1997. PMID: 9328129 Free PMC article.
-
Mutations of the von Hippel-Lindau gene confer increased susceptibility to natural killer cells of clear-cell renal cell carcinoma.Oncogene. 2011 Jun 9;30(23):2622-32. doi: 10.1038/onc.2010.638. Epub 2011 Jan 24. Oncogene. 2011. PMID: 21258414
-
Mutations in renal cell carcinoma.Urol Oncol. 2020 Oct;38(10):763-773. doi: 10.1016/j.urolonc.2018.10.027. Epub 2018 Nov 23. Urol Oncol. 2020. PMID: 30478013 Review.
-
Structure, expression and function of HLA-G in renal cell carcinoma.Semin Cancer Biol. 2007 Dec;17(6):444-50. doi: 10.1016/j.semcancer.2007.07.001. Epub 2007 Jul 13. Semin Cancer Biol. 2007. PMID: 17707652 Review.
Cited by
-
PD1/PD-L1 blockade in clear cell renal cell carcinoma: mechanistic insights, clinical efficacy, and future perspectives.Mol Cancer. 2024 Jul 16;23(1):146. doi: 10.1186/s12943-024-02059-y. Mol Cancer. 2024. PMID: 39014460 Free PMC article. Review.
-
Identification of an immune subtype-related prognostic signature of clear cell renal cell carcinoma based on single-cell sequencing analysis.Front Oncol. 2023 Mar 22;13:1067987. doi: 10.3389/fonc.2023.1067987. eCollection 2023. Front Oncol. 2023. PMID: 37035172 Free PMC article.
-
NK cells in renal cell carcinoma and its implications for CAR-NK therapy.Front Cell Dev Biol. 2025 Feb 20;13:1532491. doi: 10.3389/fcell.2025.1532491. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40052147 Free PMC article. Review.
-
Unraveling the Therapeutic Potential of Scutellarin for Clear Cell Renal Cell Carcinoma: A Comprehensive Molecular Analysis.Curr Pharm Des. 2025;31(25):2038-2062. doi: 10.2174/0113816128340451241224055536. Curr Pharm Des. 2025. PMID: 40265430
-
Molecular mechanisms of immunotherapy resistance in triple-negative breast cancer.Front Immunol. 2023 Jun 23;14:1153990. doi: 10.3389/fimmu.2023.1153990. eCollection 2023. Front Immunol. 2023. PMID: 37426654 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
