Genome-wide identification, evolution, and expression analysis of the NPR1- like gene family in pears
- PMID: 35003927
- PMCID: PMC8684321
- DOI: 10.7717/peerj.12617
Genome-wide identification, evolution, and expression analysis of the NPR1- like gene family in pears
Abstract
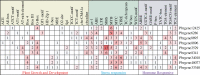
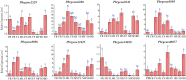
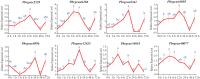
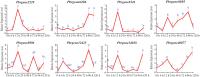
The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) plays a master regulatory role in the salicylic acid (SA) signal transduction pathway and plant systemic acquired resistance (SAR). Members of the NPR1-like gene family have been reported to the associated with biotic/abiotic stress in many plants, however the genome-wide characterization of NPR1-like genes has not been carried out in Chinese pear (Pyrus bretschneideri Reld). In this study, a systematic analysis was conducted on the characteristics of the NPR1-like genes in P. bretschneideri Reld at the whole-genome level. A total nine NPR1-like genes were detected which eight genes were located on six chromosomes and one gene was mapped to scaffold. Based on the phylogenetic analysis, the nine PbrNPR1-like proteins were divided into three clades (Clades I-III) had similar gene structure, domain and conserved motifs. We sorted the cis-acting elements into three clades, including plant growth and development, stress responses, and hormone responses in the promoter regions of PbrNPR1-like genes. The result of qPCR analysis showed that expression diversity of PbrNPR1-like genes in various tissues. All the genes were up-regulated after SA treatment in leaves except for Pbrgene8896. PbrNPR1-like genes showed circadian rhythm and significantly different expression levels after inoculation with Alternaria alternata. These findings provide a solid insight for understanding the functions and evolution of PbrNPR1-like genes in Chinese pear.
Keywords: Expression; NPR1; Pear; Phylogenetic; SAR; Salicylic acid.
© 2021 Wei et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Genome-Wide Identification and Analysis of the NPR1-Like Gene Family in Bread Wheat and Its Relatives.Int J Mol Sci. 2019 Nov 27;20(23):5974. doi: 10.3390/ijms20235974. Int J Mol Sci. 2019. PMID: 31783558 Free PMC article.
-
Genome-wide identification and characterization of bZIP transcription factors and their expression profile under abiotic stresses in Chinese pear (Pyrus bretschneideri).BMC Plant Biol. 2021 Sep 9;21(1):413. doi: 10.1186/s12870-021-03191-3. BMC Plant Biol. 2021. PMID: 34503442 Free PMC article.
-
Characterization and expression patterns of the NPR1-like genes in maize.J Genet. 2025;104:8. J Genet. 2025. PMID: 40171793
-
The NONEXPRESSOR OF PATHOGENESIS-RELATED GENES 1 (NPR1) and Related Family: Mechanistic Insights in Plant Disease Resistance.Front Plant Sci. 2019 Feb 13;10:102. doi: 10.3389/fpls.2019.00102. eCollection 2019. Front Plant Sci. 2019. PMID: 30815005 Free PMC article. Review.
-
More stories to tell: NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1, a salicylic acid receptor.Plant Cell Environ. 2021 Jun;44(6):1716-1727. doi: 10.1111/pce.14003. Epub 2021 Feb 3. Plant Cell Environ. 2021. PMID: 33495996 Review.
Cited by
-
Unraveling NPR-like Family Genes in Fragaria spp. Facilitated to Identify Putative NPR1 and NPR3/4 Orthologues Participating in Strawberry-Colletotrichum fructicola Interaction.Plants (Basel). 2022 Jun 16;11(12):1589. doi: 10.3390/plants11121589. Plants (Basel). 2022. PMID: 35736739 Free PMC article.
-
Seeing is believing: Understanding functions of NPR1 and its paralogs in plant immunity through cellular and structural analyses.Curr Opin Plant Biol. 2023 Jun;73:102352. doi: 10.1016/j.pbi.2023.102352. Epub 2023 Mar 17. Curr Opin Plant Biol. 2023. PMID: 36934653 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

