β-Tubulin Isotype, TUBB4B, Regulates The Maintenance of Cancer Stem Cells
- PMID: 35004310
- PMCID: PMC8733585
- DOI: 10.3389/fonc.2021.788024
β-Tubulin Isotype, TUBB4B, Regulates The Maintenance of Cancer Stem Cells
Abstract
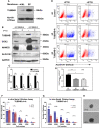
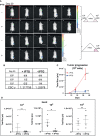
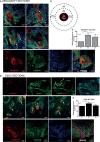
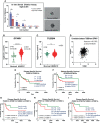
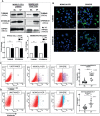
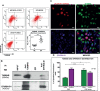
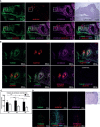
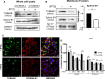
Recent advancements in cancer research have shown that cancer stem cell (CSC) niche is a crucial factor modulating tumor progression and treatment outcomes. It sustains CSCs by orchestrated regulation of several cytokines, growth factors, and signaling pathways. Although the features defining adult stem cell niches are well-explored, the CSC niche is poorly characterized. Since membrane trafficking proteins have been shown to be essential for the localization of critical proteins supporting CSCs, we investigated the role of TUBB4B, a probable membrane trafficking protein that was found to be overexpressed in the membranes of stem cell enriched cultures, in sustaining CSCs in oral cancer. Here, we show that the knockdown of TUBB4B downregulates the expression of pluripotency markers, depletes ALDH1A1+ population, decreases in vitro sphere formation, and diminishes the tumor initiation potential in vivo. As TUBB4B is not known to have any role in transcriptional regulation nor cell signaling, we suspected that its membrane trafficking function plays a role in constituting a CSC niche. The pattern of its expression in tissue sections, forming a gradient in and around the CSCs, reinforced the notion. Later, we explored its possible cooperation with a signaling protein, Ephrin-B1, the abrogation of which reduces the self-renewal of oral cancer stem cells. Expression and survival analyses based on the TCGA dataset of head and neck squamous cell carcinoma (HNSCC) samples indicated that the functional cooperation of TUBB4 and EFNB1 results in a poor prognosis. We also show that TUBB4B and Ephrin-B1 cohabit in the CSC niche. Moreover, depletion of TUBB4B downregulates the membrane expression of Ephrin-B1 and reduces the CSC population. Our results imply that the dynamics of TUBB4B is decisive for the surface localization of proteins, like Ephrin-B1, that sustain CSCs by their concerted signaling.
Keywords: ALDH1A1; CSC niche; TUBB4B (Tubulin β-2C); cancer stem cells; ephrin-B1.
Copyright © 2021 Dharmapal, Jyothy, Mohan, Balagopal, George, Sebastian, Maliekal and Sengupta.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Tubulin Isotypes: Emerging Roles in Defining Cancer Stem Cell Niche.Front Immunol. 2022 May 26;13:876278. doi: 10.3389/fimmu.2022.876278. eCollection 2022. Front Immunol. 2022. PMID: 35693789 Free PMC article. Review.
-
Glutathione metabolism is essential for self-renewal and chemoresistance of pancreatic cancer stem cells.World J Stem Cells. 2020 Nov 26;12(11):1410-1428. doi: 10.4252/wjsc.v12.i11.1410. World J Stem Cells. 2020. PMID: 33312407 Free PMC article.
-
SMURF1 silencing diminishes a CD44-high cancer stem cell-like population in head and neck squamous cell carcinoma.Mol Cancer. 2014 Dec 3;13:260. doi: 10.1186/1476-4598-13-260. Mol Cancer. 2014. PMID: 25471937 Free PMC article.
-
Cancer stem cells enrichment with surface markers CD271 and CD44 in human head and neck squamous cell carcinomas.Carcinogenesis. 2020 Jun 17;41(4):458-466. doi: 10.1093/carcin/bgz182. Carcinogenesis. 2020. PMID: 31742606 Free PMC article.
-
Cancer stem cells and their niche in the progression of squamous cell carcinoma.Cancer Sci. 2020 Nov;111(11):3985-3992. doi: 10.1111/cas.14639. Epub 2020 Sep 18. Cancer Sci. 2020. PMID: 32888236 Free PMC article. Review.
Cited by
-
Two-Dimensional-PAGE Coupled with nLC-MS/MS-Based Identification of Differentially Expressed Proteins and Tumorigenic Pathways in MCF7 Breast Cancer Cells Transfected for JTB Protein Silencing.Molecules. 2023 Nov 9;28(22):7501. doi: 10.3390/molecules28227501. Molecules. 2023. PMID: 38005222 Free PMC article.
-
Hindbrain boundaries as niches of neural progenitor and stem cells regulated by the extracellular matrix proteoglycan chondroitin sulphate.Development. 2024 Feb 15;151(4):dev201934. doi: 10.1242/dev.201934. Epub 2024 Feb 13. Development. 2024. PMID: 38251863 Free PMC article.
-
Exploring the complex role of the Eph/Ephrin signaling in oral and maxillofacial cancers.Front Oncol. 2025 May 12;15:1554751. doi: 10.3389/fonc.2025.1554751. eCollection 2025. Front Oncol. 2025. PMID: 40421081 Free PMC article. Review.
-
Metabolic shifts in glioblastoma: unraveling altered pathways and exploring novel therapeutic avenues.Mol Biol Rep. 2025 Jan 22;52(1):146. doi: 10.1007/s11033-025-10242-7. Mol Biol Rep. 2025. PMID: 39841290 Review.
-
Tubulin Isotypes: Emerging Roles in Defining Cancer Stem Cell Niche.Front Immunol. 2022 May 26;13:876278. doi: 10.3389/fimmu.2022.876278. eCollection 2022. Front Immunol. 2022. PMID: 35693789 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

