Deciphering the Mechanism of Gilteritinib Overcoming Lorlatinib Resistance to the Double Mutant I1171N/F1174I in Anaplastic Lymphoma Kinase
- PMID: 35004700
- PMCID: PMC8733690
- DOI: 10.3389/fcell.2021.808864
Deciphering the Mechanism of Gilteritinib Overcoming Lorlatinib Resistance to the Double Mutant I1171N/F1174I in Anaplastic Lymphoma Kinase
Abstract
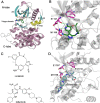
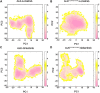
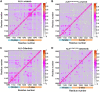
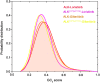
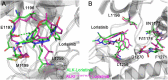
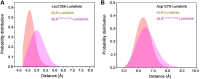
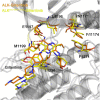
Anaplastic lymphoma kinase (ALK) is validated as a therapeutic molecular target in multiple malignancies, such as non-small cell lung cancer (NSCLC). However, the feasibility of targeted therapies exerted by ALK inhibitors is inevitably hindered owing to drug resistance. The emergence of clinically acquired drug mutations has become a major challenge to targeted therapies and personalized medicines. Thus, elucidating the mechanism of resistance to ALK inhibitors is helpful for providing new therapeutic strategies for the design of next-generation drug. Here, we used molecular docking and multiple molecular dynamics simulations combined with correlated and energetical analyses to explore the mechanism of how gilteritinib overcomes lorlatinib resistance to the double mutant ALK I1171N/F1174I. We found that the conformational dynamics of the ALK kinase domain was reduced by the double mutations I1171N/F1174I. Moreover, energetical and structural analyses implied that the double mutations largely disturbed the conserved hydrogen bonding interactions from the hinge residues Glu1197 and Met1199 in the lorlatinib-bound state, whereas they had no discernible adverse impact on the binding affinity and stability of gilteritinib-bound state. These discrepancies created the capacity of the double mutant ALK I1171N/F1174I to confer drug resistance to lorlatinib. Our result anticipates to provide a mechanistic insight into the mechanism of drug resistance induced by ALK I1171N/F1174I that are resistant to lorlatinib treatment in NSCLC.
Keywords: anaplastic lymphoma kinase; drug resistance; molecular dynamics simulations; non-small cell lung cancer; targeted therapy.
Copyright © 2021 Liang, Wang, Qi, Liu, Li, Lu, Mou and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., et al. (2015). Gromacs: High Performance Molecular Simulations through Multi-Level Parallelism from Laptops to Supercomputers. SoftwareX 1-2, 19–25. 10.1016/j.softx.2015.06.001 - DOI
-
- Anggayasti W. L., Ogino K., Yamamoto E., Helmerhorst E., Yasuoka K., Mancera R. L. (2020). The Acidic Tail of HMGB1 Regulates its Secondary Structure and Conformational Flexibility: A Circular Dichroism and Molecular Dynamics Simulation Study. Comput. Struct. Biotechnol. J. 18, 1160–1172. 10.1016/j.csbj.2020.05.012 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

