Transcriptomic Responses of Cordyceps militaris to Salt Treatment During Cordycepins Production
- PMID: 35004818
- PMCID: PMC8733472
- DOI: 10.3389/fnut.2021.793795
Transcriptomic Responses of Cordyceps militaris to Salt Treatment During Cordycepins Production
Abstract
Cordycepin is a major bioactive compound found in Cordyceps militaris (C. militaris) that exhibits a broad spectrum of biological activities. Hence, it is potentially a bioactive ingredient of pharmaceutical and cosmetic products. However, overexploitation and low productivity of natural C. militaris is a barrier to commercialization, which leads to insufficient supply to meet its existing market demands. In this study, a preliminary study of distinct concentrations of salt treatments toward C. militaris was conducted. Although the growth of C. militaris was inhibited by different salt treatments, the cordycepin production increased significantly accompanied by the increment of salt concentration. Among them, the content of cordycepin in the 7% salt-treated group was five-fold higher than that of the control group. Further transcriptome analysis of samples with four salt concentrations, coupled with Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, several differentially expressed genes (DEGs) were found. Finally, dynamic changes of the expression patterns of four genes involved in the cordycepin biosynthesis pathway were observed by the quantitative real-time PCR. Taken together, our study provides a global transcriptome characterization of the salt treatment adaptation process in C. militaris and facilitates the construction of industrial strains with a high cordycepin production and salt tolerance.
Keywords: Cordyceps militaris; cordycepin; molecular mechanism; salt treatment; transcriptome.
Copyright © 2021 Lv, Zhu, Cheng, Cao, Zeng, Liu and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

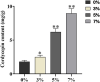




References
-
- Wang L, Zhang WM, Hu B, Chen YQ, Qu LH. Genetic variation of Cordyceps militaris and its allies based on phylogenetic analysis of rDNA ITS sequence data. Fungal Divers. (2008) 31:147–55. Available online at: https://www.fungaldiversity.org/fdp/sfdp/31-11.pdf
-
- Shrestha B, Tanaka E, Hyun MW, Han J-G, Kim CS, Jo JW, et al. . Coleopteran and lepidopteran hosts of the entomopathogenic genus cordyceps sensu lato. J Mycol. (2016) 2016:1–14. 10.1155/2016/7648219 - DOI
LinkOut - more resources
Full Text Sources

