Energy Bilocalization Effect and the Emergence of Molecular Functions in Proteins
- PMID: 35004841
- PMCID: PMC8733615
- DOI: 10.3389/fmolb.2021.736376
Energy Bilocalization Effect and the Emergence of Molecular Functions in Proteins
Abstract
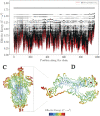
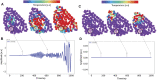
Proteins are among the most complex molecular structures, which have evolved to develop broad functions, such as energy conversion and transport, information storage and processing, communication, and regulation of chemical reactions. However, the mechanisms by which these dynamical entities coordinate themselves to perform biological tasks remain hotly debated. Here, a physical theory is presented to explain how functional dynamical behavior possibly emerge in complex/macro molecules, thanks to the effect that we term bilocalization of thermal vibrations. More specifically, our approach allows us to understand how structural irregularities lead to a partitioning of the energy of the vibrations into two distinct sets of molecular domains, corresponding to slow and fast motions. This shape-encoded spectral allocation, associated to the genetic sequence, provides a close access to a wide reservoir of dynamical patterns, and eventually allows the emergence of biological functions by natural selection. To illustrate our approach, the SPIKE protein structure of SARS-COV2 is considered.
Keywords: dynamics; localization; protein; rate promoting vibrations; vibrations.
Copyright © 2021 Chalopin and Sparfel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- Anderson P. W. (1958). Absence of Diffusion in Certain Random Lattices. Phys. Rev. 109, 1492–1505. 10.1103/physrev.109.1492 - DOI
-
- Antoniou D., Schwartz S. D. (2001). Internal Enzyme Motions as a Source of Catalytic Activity: Rate-Promoting Vibrations and Hydrogen Tunneling. J. Phys. Chem. B 105, 5553–5558. 10.1021/jp004547b - DOI
-
- Ashcroft Neil. W. (1976). in Solid State Physics/Neil W. Ashcroft. Editor David Mermin N. (Philadelphia (Pa.): Saunders College; ).
LinkOut - more resources
Full Text Sources
Miscellaneous

