Sequential Coupling Shows Minor Effects of Fluid Dynamics on Myocardial Deformation in a Realistic Whole-Heart Model
- PMID: 35004885
- PMCID: PMC8733159
- DOI: 10.3389/fcvm.2021.768548
Sequential Coupling Shows Minor Effects of Fluid Dynamics on Myocardial Deformation in a Realistic Whole-Heart Model
Abstract
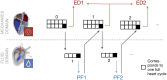
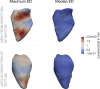
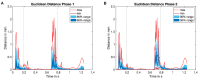
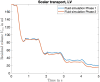
Background: The human heart is a masterpiece of the highest complexity coordinating multi-physics aspects on a multi-scale range. Thus, modeling the cardiac function in silico to reproduce physiological characteristics and diseases remains challenging. Especially the complex simulation of the blood's hemodynamics and its interaction with the myocardial tissue requires a high accuracy of the underlying computational models and solvers. These demanding aspects make whole-heart fully-coupled simulations computationally highly expensive and call for simpler but still accurate models. While the mechanical deformation during the heart cycle drives the blood flow, less is known about the feedback of the blood flow onto the myocardial tissue. Methods and Results: To solve the fluid-structure interaction problem, we suggest a cycle-to-cycle coupling of the structural deformation and the fluid dynamics. In a first step, the displacement of the endocardial wall in the mechanical simulation serves as a unidirectional boundary condition for the fluid simulation. After a complete heart cycle of fluid simulation, a spatially resolved pressure factor (PF) is extracted and returned to the next iteration of the solid mechanical simulation, closing the loop of the iterative coupling procedure. All simulations were performed on an individualized whole heart geometry. The effect of the sequential coupling was assessed by global measures such as the change in deformation and-as an example of diagnostically relevant information-the particle residence time. The mechanical displacement was up to 2 mm after the first iteration. In the second iteration, the deviation was in the sub-millimeter range, implying that already one iteration of the proposed cycle-to-cycle coupling is sufficient to converge to a coupled limit cycle. Conclusion: Cycle-to-cycle coupling between cardiac mechanics and fluid dynamics can be a promising approach to account for fluid-structure interaction with low computational effort. In an individualized healthy whole-heart model, one iteration sufficed to obtain converged and physiologically plausible results.
Keywords: cardiovascular modeling; fluid dynamics simulation; fluid-structure interaction; hemodynamics; multi-physics coupling; whole heart.
Copyright © 2021 Brenneisen, Daub, Gerach, Kovacheva, Huetter, Frohnapfel, Dössel and Loewe.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Miscellaneous

