Intracellular IL-32 regulates mitochondrial metabolism, proliferation, and differentiation of malignant plasma cells
- PMID: 35005550
- PMCID: PMC8717606
- DOI: 10.1016/j.isci.2021.103605
Intracellular IL-32 regulates mitochondrial metabolism, proliferation, and differentiation of malignant plasma cells
Abstract
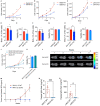
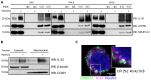
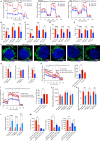
Interleukin-32 (IL-32) is a nonclassical cytokine expressed in cancers, inflammatory diseases, and infections. Its expression is regulated by two different oxygen sensing systems; HIF1α and cysteamine dioxygenase (ADO), indicating that IL-32 may be involved in the response to hypoxia. We here demonstrate that endogenously expressed, intracellular IL-32 interacts with components of the mitochondrial respiratory chain and promotes oxidative phosphorylation. Knocking out IL-32 in three myeloma cell lines reduced cell survival and proliferation in vitro and in vivo. High-throughput transcriptomic and MS-metabolomic profiling of IL-32 KO cells revealed that cells depleted of IL-32 had perturbations in metabolic pathways, with accumulation of lipids, pyruvate precursors, and citrate. IL-32 was expressed in a subgroup of myeloma patients with inferior survival, and primary myeloma cells expressing IL-32 had a gene signature associated with immaturity, proliferation, and oxidative phosphorylation. In conclusion, we demonstrate a previously unrecognized role of IL-32 in the regulation of plasma cell metabolism.
Keywords: Cancer; Cell biology; Immunology.
© 2021 The Author(s).
Conflict of interest statement
There are no conflicts of interests.
Figures



References
-
- Azab A.K., Hu J., Quang P., Azab F., Pitsillides C., Awwad R., Thompson B., Maiso P., Sun J.D., Hart C.P., et al. Hypoxia promotes dissemination of multiple myeloma through acquisition of epithelial to mesenchymal transition-like features. Blood. 2012;119:5782–5794. doi: 10.1182/blood-2011-09-380410. - DOI - PMC - PubMed
-
- Bailur J.K., McCachren S.S., Doxie D.B., Shrestha M., Pendleton K., Nooka A.K., Neparidze N., Parker T.L., Bar N., Kaufman J.L., et al. Early alterations in stem-like/marrow-resident T cells and innate and myeloid cells in preneoplastic gammopathy. JCI Insight. 2019;4:e127807. doi: 10.1172/jci.insight.127807. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

