RQC helical hairpin in Bloom's syndrome helicase regulates DNA unwinding by dynamically intercepting nascent nucleotides
- PMID: 35005551
- PMCID: PMC8718986
- DOI: 10.1016/j.isci.2021.103606
RQC helical hairpin in Bloom's syndrome helicase regulates DNA unwinding by dynamically intercepting nascent nucleotides
Abstract
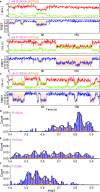
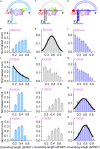
The RecQ family of helicases are important for maintenance of genomic integrity. Although functions of constructive subdomains of this family of helicases have been extensively studied, the helical hairpin (HH) in the RecQ-C-terminal domain (RQC) has been underappreciated and remains poorly understood. Here by using single-molecule fluorescence resonance energy transfer, we found that HH in the human BLM transiently intercepts different numbers of nucleotides when it is unwinding a double-stranded DNA. Single-site mutations in HH that disrupt hydrogen bonds and/or salt bridges between DNA and HH change the DNA binding conformations and the unwinding features significantly. Our results, together with recent clinical tests that correlate single-site mutations in HH of human BLM with the phenotype of cancer-predisposing syndrome or Bloom's syndrome, implicate pivotal roles of HH in BLM's DNA unwinding activity. Similar mechanisms might also apply to other RecQ family helicases, calling for more attention to the RQC helical hairpin.
Keywords: Analytical chemistry instrumentation; Biochemistry; Biological sciences; Structural biology.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Structural mechanisms of human RecQ helicases WRN and BLM.Front Genet. 2014 Oct 29;5:366. doi: 10.3389/fgene.2014.00366. eCollection 2014. Front Genet. 2014. PMID: 25400656 Free PMC article. Review.
-
Structure of human Bloom's syndrome helicase in complex with ADP and duplex DNA.Acta Crystallogr D Biol Crystallogr. 2014 May;70(Pt 5):1465-75. doi: 10.1107/S139900471400501X. Epub 2014 Apr 30. Acta Crystallogr D Biol Crystallogr. 2014. PMID: 24816114
-
Unwinding forward and sliding back: an intermittent unwinding mode of the BLM helicase.Nucleic Acids Res. 2015 Apr 20;43(7):3736-46. doi: 10.1093/nar/gkv209. Epub 2015 Mar 12. Nucleic Acids Res. 2015. PMID: 25765643 Free PMC article.
-
The Bloom's syndrome helicase unwinds G4 DNA.J Biol Chem. 1998 Oct 16;273(42):27587-92. doi: 10.1074/jbc.273.42.27587. J Biol Chem. 1998. PMID: 9765292
-
Bloom's syndrome: Why not premature aging?: A comparison of the BLM and WRN helicases.Ageing Res Rev. 2017 Jan;33:36-51. doi: 10.1016/j.arr.2016.05.010. Epub 2016 May 26. Ageing Res Rev. 2017. PMID: 27238185 Free PMC article. Review.
Cited by
-
Mesoscale DNA features impact APOBEC3A and APOBEC3B deaminase activity and shape tumor mutational landscapes.Nat Commun. 2024 Mar 18;15(1):2370. doi: 10.1038/s41467-024-45909-5. Nat Commun. 2024. PMID: 38499542 Free PMC article.

