The evolution of knowledge on genes associated with human diseases
- PMID: 35005554
- PMCID: PMC8719018
- DOI: 10.1016/j.isci.2021.103610
The evolution of knowledge on genes associated with human diseases
Abstract
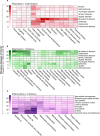
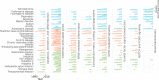
Thousands of biomedical scientific articles, including those describing genes associated with human diseases, are published every week. Computational methods such as text mining and machine learning algorithms are now able to automatically detect these associations. In this study, we used a cognitive computing text-mining application to construct a knowledge network comprising 3,723 genes and 99 diseases. We then tracked the yearly changes on these networks to analyze how our knowledge has evolved in the past 30 years. Our systems approach helped to unravel the molecular bases of diseases and detect shared mechanisms between clinically distinct diseases. It also revealed that multi-purpose therapeutic drugs target genes that are commonly associated with several psychiatric, inflammatory, or infectious disorders. By navigating this knowledge tsunami, we were able to extract relevant biological information and insights about human diseases.
Keywords: Association analysis; Bioinformatics; Molecular network; Systems biology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- de Baumont A., Maschietto M., Lima L., Carraro D.M., Olivieri E.H., Fiorini A., Barreta L.A.N., Palha J.A., Belmonte-de-Abreu P., Moreira Filho C.A., Brentani H. Innate immune response is differentially dysregulated between bipolar disease and schizophrenia. Schizophr. Res. 2015;161:215–221. doi: 10.1016/j.schres.2014.10.055. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

