Gut Ruminococcaceae levels at baseline correlate with risk of antibiotic-associated diarrhea
- PMID: 35005566
- PMCID: PMC8718891
- DOI: 10.1016/j.isci.2021.103644
Gut Ruminococcaceae levels at baseline correlate with risk of antibiotic-associated diarrhea
Abstract
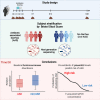
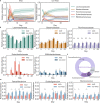
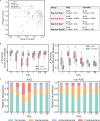
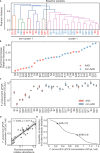
Antibiotic-associated diarrhea (AAD) affects a significant proportion of patients receiving antibiotics. We sought to understand if differences in the gut microbiome would influence the development of AAD. We administered a 3-day course of amoxicillin-clavulanate to 30 healthy adult volunteers, and analyzed their stool microbiome, using 16S rRNA gene sequencing, at baseline and up to 4 weeks post antibiotic administration. Lower levels of gut Ruminococcaceae were significantly and consistently observed from baseline until day 7 in participants who developed AAD. Overall, participants who developed AAD experienced a greater decrease in microbial diversity. The probability of AAD could be predicted based on qPCR-derived levels of Faecalibacterium prausnitzii at baseline. Our findings suggest that a lack of gut Ruminococcaceae influences development of AAD. Quantification of F. prausnitzii in stool prior to antibiotic administration may help identify patients at risk of AAD, and aid clinicians in devising individualized treatment regimens to minimize such adverse effects.
Keywords: Health sciences; Microbiome; Pathophysiology.
© 2021 The Author(s).
Conflict of interest statement
Eric J Alm is co-founder of Finch Therapeutics, a microbiome-based therapeutics company. All other authors have no conflicts of interests to declare.
Figures

References
-
- Allen S.J., Wareham K., Wang D., Bradley C., Hutchings H., Harris W., Dhar A., Brown H., Foden A., Gravenor M.B., et al. Lactobacilli and bifidobacteria in the prevention of antibiotic-associated diarrhoea and Clostridium difficile diarrhoea in older inpatients (PLACIDE): a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2013;382:1249–1257. - PubMed
-
- Bartlett J.G. Antibiotic-associated diarrhea. Clin. Infect. Dis. 2015;18:349–351. - PubMed
-
- Beaugerie L., Petit J.C. Antibiotic-associated diarrhoea. Best Pract. Res. Clin. Gastroenterol. 2004;18:337–352. - PubMed
LinkOut - more resources
Full Text Sources

