Abnormal sialylation and fucosylation of saliva glycoproteins: Characteristics of lung cancer-specific biomarkers
- PMID: 35005612
- PMCID: PMC8718573
- DOI: 10.1016/j.crphar.2021.100079
Abnormal sialylation and fucosylation of saliva glycoproteins: Characteristics of lung cancer-specific biomarkers
Abstract
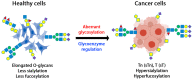
Dysregulated surface glycoproteins play an important role in tumor cell proliferation and progression. Abnormal glycosylation of these glycoproteins may activate tumor signal transduction and lead to tumor development. The tumor microenvironment alters its molecular composition, some of which regulate protein glycosylation biosynthesis. The glycosylation of saliva proteins in lung cancer patients is different from healthy controls, in which the glycans of cancer patients are highly sialylated and hyperfucosylated. Most studies have shown that O-glycans from cancer are truncated O-glycans, while N-glycans contain fucoses and sialic acids. Because glycosylation analysis is challenging, there are few reports on how glycosylation of saliva proteins is related to the occurrence or progression of lung cancer. In this review, we discussed glycoenzymes involved in protein glycosylation, their changes in tumor microenvironment, potential tumor biomarkers present in body fluids, and abnormal glycosylation of saliva or lung glycoproteins. We further explored the effect of glycosylation changes on tumor signal transduction, and emphasized the role of receptor tyrosine kinases in tumorigenesis and metastasis.
Keywords: Glycoenzyme; Glycosylation; Mass spectrometry; Saliva; Tumor biomarker.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing financial interests.
Figures



Similar articles
-
Quantitative analysis of fucosylated glycoproteins by immobilized lectin-affinity fluorescent labeling.RSC Adv. 2023 Feb 27;13(10):6676-6687. doi: 10.1039/d3ra00072a. eCollection 2023 Feb 21. RSC Adv. 2023. PMID: 36860533 Free PMC article.
-
Aberrant Fucosylation of Saliva Glycoprotein Defining Lung Adenocarcinomas Malignancy.ACS Omega. 2022 May 19;7(21):17894-17906. doi: 10.1021/acsomega.2c01193. eCollection 2022 May 31. ACS Omega. 2022. PMID: 35664632 Free PMC article.
-
Role of tumor cell sialylation in pancreatic cancer progression.Adv Cancer Res. 2023;157:123-155. doi: 10.1016/bs.acr.2022.07.003. Epub 2022 Sep 27. Adv Cancer Res. 2023. PMID: 36725107 Free PMC article. Review.
-
Glycosylation characteristics of colorectal cancer.Adv Cancer Res. 2015;126:203-56. doi: 10.1016/bs.acr.2014.11.004. Epub 2015 Feb 7. Adv Cancer Res. 2015. PMID: 25727149 Review.
-
Targeting aberrant sialylation and fucosylation in prostate cancer cells using potent metabolic inhibitors.Glycobiology. 2023 Dec 30;33(12):1155-1171. doi: 10.1093/glycob/cwad085. Glycobiology. 2023. PMID: 37847613 Free PMC article.
Cited by
-
Identification of a potential sialylation-related pattern for the prediction of prognosis and immunotherapy response in small cell lung cancer.Medicine (Baltimore). 2024 Oct 11;103(41):e40060. doi: 10.1097/MD.0000000000040060. Medicine (Baltimore). 2024. PMID: 39465788 Free PMC article.
-
Quantitative analysis of fucosylated glycoproteins by immobilized lectin-affinity fluorescent labeling.RSC Adv. 2023 Feb 27;13(10):6676-6687. doi: 10.1039/d3ra00072a. eCollection 2023 Feb 21. RSC Adv. 2023. PMID: 36860533 Free PMC article.
-
Morphology and Glycan Composition of the Mandibular Glands in the White-Eared Opossum (Didelphis albiventris).J Morphol. 2025 Aug;286(8):e70074. doi: 10.1002/jmor.70074. J Morphol. 2025. PMID: 40817713 Free PMC article.
-
Advances in the Immunomodulatory Properties of Glycoantigens in Cancer.Cancers (Basel). 2022 Apr 7;14(8):1854. doi: 10.3390/cancers14081854. Cancers (Basel). 2022. PMID: 35454762 Free PMC article. Review.
-
Targeting protein glycosylation to regulate inflammation in the respiratory tract: novel diagnostic and therapeutic candidates for chronic respiratory diseases.Front Immunol. 2023 May 15;14:1168023. doi: 10.3389/fimmu.2023.1168023. eCollection 2023. Front Immunol. 2023. PMID: 37256139 Free PMC article. Review.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

