Mapping atypical UV photoproducts in vitro and across the S. cerevisiae genome
- PMID: 35005641
- PMCID: PMC8715331
- DOI: 10.1016/j.xpro.2021.101059
Mapping atypical UV photoproducts in vitro and across the S. cerevisiae genome
Abstract
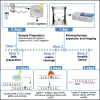
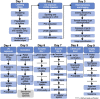
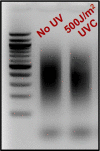
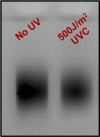
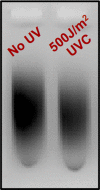
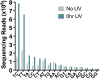
Exposure to ultraviolet (UV) light induces DNA damage, predominantly cyclobutane pyrimidine dimers (CPD) and 6,4-photoproducts (6,4-PP), as well as rare, atypical photoproducts at thymidine-adenine (TA) sequences. We have recently shown 'TA' photoproducts are induced in UV-irradiated oligonucleotides and across the budding yeast genome. Here, we describe a protocol for mapping atypical 'TA' photoproducts in vitro and in vivo. This protocol overcomes the technical challenges involved in accurately mapping such rare photoproducts by using ultraviolet damage endonuclease (UVDE) enzymes. For complete details on the use and execution of this protocol, please refer to Laughery et al. (2020).
Keywords: Bioinformatics; Model Organisms; Molecular Biology; Sequence analysis; Sequencing.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

