Binding of AP endonuclease-1 to G-quadruplex DNA depends on the N-terminal domain, Mg2+ and ionic strength
- PMID: 35005714
- PMCID: PMC8740889
- DOI: 10.1021/acsbiomedchemau.1c00031
Binding of AP endonuclease-1 to G-quadruplex DNA depends on the N-terminal domain, Mg2+ and ionic strength
Abstract
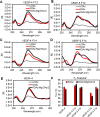
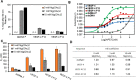
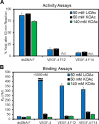
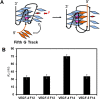
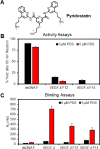
The base excision repair enzyme apurinic/apyrimidinic endonuclease-1 (APE1) is also engaged in transcriptional regulation. APE1 can function in both pathways when the protein binds to a promoter G-quadruplex (G4) bearing an abasic site (modeled with tetrahydrofuran, F) that leads to enzymatic stalling on the non-canonical fold to recruit activating transcription factors. Biochemical and biophysical studies to address APE1's binding and catalytic activity with the vascular endothelial growth factor (VEGF) promoter G4 are lacking, and the present work provides insight on this topic. Herein, the native APE1 was used for cleavage assays, and the catalytically inactive mutant D210A was used for binding assays with double-stranded DNA (dsDNA) versus the native G4 or the G4 with F at various positions, revealing dependencies of the interaction on the cation concentrations K+ and Mg2+ and the N-terminal domain of the protein. Assays in 0, 1, or 10 mM Mg2+ found that dsDNA and G4 substrates required the cation for both binding and catalysis, in which G4 binding increased with [Mg2+]. Studies with 50 versus physiological 140 mM K+ ions showed that F-containing dsDNA was bound and cleaved by APE1; whereas, the G4s with F were poorly cleaved in low salt and not cleaved at all at higher salt while the binding remained robust. Using Δ33 or Δ61 N-terminal truncated APE1 proteins, the binding and cleavage of dsDNA with F was minimally impacted; in contrast, the G4s required the N-terminus for binding and catalysis is nearly abolished without the N-terminus. With this knowledge, we found APE1 could remodel the F-containing VEGF promoter dsDNA→G4 folds in solution. Lastly, the addition of the G4 ligand pyridostatin inhibited APE1 binding and cleavage of F-containing G4s but not dsDNA. The biological and medicinal chemistry implications of the results are discussed.
Keywords: APE1; DNA remodeling; G-Quadruplex; binding assays; gene regulation; pyridostatin.
Conflict of interest statement
Conflict of Interest. No conflicts of interest are declared in this work.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

