Transcriptional response of Wolbachia-transinfected Aedes aegypti mosquito cells to dengue virus at early stages of infection
- PMID: 35006065
- PMCID: PMC8895618
- DOI: 10.1099/jgv.0.001694
Transcriptional response of Wolbachia-transinfected Aedes aegypti mosquito cells to dengue virus at early stages of infection
Abstract
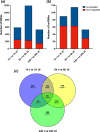
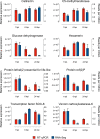
Mosquito-borne flaviviruses are responsible for viral infections and represent a considerable public health burden. Aedes aegypti is the principal vector of dengue virus (DENV), therefore understanding the intrinsic virus-host interactions is vital, particularly in the presence of the endosymbiont Wolbachia, which blocks virus replication in mosquitoes. Here, we examined the transcriptional response of Wolbachia-transinfected Ae. aegypti Aag2 cells to DENV infection. We identified differentially expressed immune genes that play a key role in the activation of anti-viral defence such as the Toll and immune deficiency pathways. Further, genes encoding cytosine and N6-adenosine methyltransferases and SUMOylation, involved in post-transcriptional modifications, an antioxidant enzyme, and heat-shock response were up-regulated at the early stages of DENV infection and are reported here for the first time. Additionally, several long non-coding RNAs were among the differentially regulated genes. Our results provide insight into Wolbachia-transinfected Ae. aegypti's initial virus recognition and transcriptional response to DENV infection.
Keywords: Aedes aegypti; Wolbachia; dengue virus; differential gene expression; transcriptome.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
-
- Liu-Helmersson J, Brännström Å, Sewe MO, Semenza JC, Rocklöv J. Estimating past, present, and future trends in the global distribution and abundance of the arbovirus vector Aedes aegypti under climate change scenarios. Front Public Health. 2019;7:148. doi: 10.3389/fpubh.2019.00148. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

