Comparative transcriptomic analysis reveals translationally relevant processes in mouse models of malaria
- PMID: 35006075
- PMCID: PMC8747512
- DOI: 10.7554/eLife.70763
Comparative transcriptomic analysis reveals translationally relevant processes in mouse models of malaria
Abstract
Recent initiatives to improve translation of findings from animal models to human disease have focussed on reproducibility but quantifying the relevance of animal models remains a challenge. Here, we use comparative transcriptomics of blood to evaluate the systemic host response and its concordance between humans with different clinical manifestations of malaria and five commonly used mouse models. Plasmodium yoelii 17XL infection of mice most closely reproduces the profile of gene expression changes seen in the major human severe malaria syndromes, accompanied by high parasite biomass, severe anemia, hyperlactatemia, and cerebral microvascular pathology. However, there is also considerable discordance of changes in gene expression between the different host species and across all models, indicating that the relevance of biological mechanisms of interest in each model should be assessed before conducting experiments. These data will aid the selection of appropriate models for translational malaria research, and the approach is generalizable to other disease models.
Keywords: genetics; genomics; malaria; mouse; mouse models; transcriptomics.
© 2022, Georgiadou et al.
Conflict of interest statement
AG, CD, PS, HL, MK, AC No competing interests declared
Figures
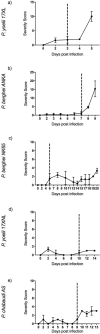

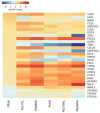
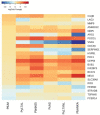

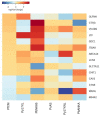
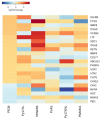
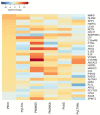
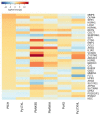

Similar articles
-
Signatures of malaria-associated pathology revealed by high-resolution whole-blood transcriptomics in a rodent model of malaria.Sci Rep. 2017 Feb 3;7:41722. doi: 10.1038/srep41722. Sci Rep. 2017. PMID: 28155887 Free PMC article.
-
Comparative histopathology of mice infected with the 17XL and 17XNL strains of Plasmodium yoelii.J Parasitol. 2012 Apr;98(2):310-5. doi: 10.1645/GE-2825.1. Epub 2011 Oct 21. J Parasitol. 2012. PMID: 22017443
-
Transcriptomic Studies of Malaria: a Paradigm for Investigation of Systemic Host-Pathogen Interactions.Microbiol Mol Biol Rev. 2018 Apr 25;82(2):e00071-17. doi: 10.1128/MMBR.00071-17. Print 2018 Jun. Microbiol Mol Biol Rev. 2018. PMID: 29695497 Free PMC article. Review.
-
Comparative Heterochromatin Profiling Reveals Conserved and Unique Epigenome Signatures Linked to Adaptation and Development of Malaria Parasites.Cell Host Microbe. 2018 Mar 14;23(3):407-420.e8. doi: 10.1016/j.chom.2018.01.008. Epub 2018 Mar 1. Cell Host Microbe. 2018. PMID: 29503181 Free PMC article.
-
Malaria pathogenesis.Science. 1994 Jun 24;264(5167):1878-83. doi: 10.1126/science.8009217. Science. 1994. PMID: 8009217 Review.
Cited by
-
A single workflow for multi-species blood transcriptomics.BMC Genomics. 2024 Mar 16;25(1):282. doi: 10.1186/s12864-024-10208-2. BMC Genomics. 2024. PMID: 38493105 Free PMC article.
-
Gene expression analyses reveal differences in children's response to malaria according to their age.Nat Commun. 2024 Mar 6;15(1):2021. doi: 10.1038/s41467-024-46416-3. Nat Commun. 2024. PMID: 38448421 Free PMC article.
-
Pathogenetic mechanisms and treatment targets in cerebral malaria.Nat Rev Neurol. 2023 Nov;19(11):688-709. doi: 10.1038/s41582-023-00881-4. Epub 2023 Oct 19. Nat Rev Neurol. 2023. PMID: 37857843 Review.
-
Effect of Chronic Stress on Whole Blood Transcriptome: A Meta-Analysis of Publicly Available Datasets from Rodent Models.bioRxiv [Preprint]. 2025 Jun 1:2025.05.30.657043. doi: 10.1101/2025.05.30.657043. bioRxiv. 2025. PMID: 40661584 Free PMC article. Preprint.
-
Comparative transcriptomics of aphid species that diverged > 22 MYA reveals genes that are important for the maintenance of their symbiosis.Sci Rep. 2023 Apr 1;13(1):5341. doi: 10.1038/s41598-023-32291-3. Sci Rep. 2023. PMID: 37005434 Free PMC article.
References
-
- Akkaya M, Bansal A, Sheehan PW, Pena M, Cimperman CK, Qi CF, Yazew T, Otto TD, Billker O, Miller LH, Pierce SK. Testing the impact of a single nucleotide polymorphism in a Plasmodium berghei ApiAP2 transcription factor on experimental cerebral malaria in mice. Scientific Reports. 2020;10:13630. doi: 10.1038/s41598-020-70617-7. - DOI - PMC - PubMed
-
- Allantaz F, Cheng DT, Bergauer T, Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H, D’Asaro M, Chiappe A, Sridhar S, Pacheco GD, Burczynski ME, Hochstrasser D, Vonderscher J, Matthes T. Expression profiling of human immune cell subsets identifies miRNA-mRNA regulatory relationships correlated with cell type specific expression. PLOS ONE. 2012;7:e29979. doi: 10.1371/journal.pone.0029979. - DOI - PMC - PubMed
-
- Amani V, Vigário AM, Belnoue E, Marussig M, Fonseca L, Mazier D, Rénia L. Involvement of IFN-gamma receptor-medicated signaling in pathology and anti-malarial immunity induced by Plasmodium berghei infection. European Journal of Immunology. 2000;30:1646–1655. doi: 10.1002/1521-4141(200006)30:6<1646::AID-IMMU1646>3.0.CO;2-0. - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

