Natural variant frequencies across domains from different sarcomere proteins cross-correlate to identify inter-protein contacts associated with cardiac muscle function and disease
- PMID: 35006463
- PMCID: PMC8607394
- DOI: 10.1186/s43556-021-00056-x
Natural variant frequencies across domains from different sarcomere proteins cross-correlate to identify inter-protein contacts associated with cardiac muscle function and disease
Abstract
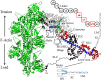
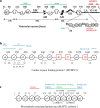
Coordinated sarcomere proteins produce contraction force for muscle shortening. In human ventriculum they include the cardiac myosin motor (βmys), repetitively converting ATP free energy into work, and myosin binding protein C (MYBPC3) that in complex with βmys is regulatory. Single nucleotide variants (SNVs) causing hereditary heart diseases frequently target this protein pair. The βmys/MYBPC3 complex models a regulated motor and is used here to study how the proteins couple. SNVs in βmys or MYBPC3 survey human populations worldwide. Their protein expression modifies domain structure affecting phenotype and pathogenicity outcomes. When the SNV modified domain locates to inter-protein contacts it could affect complex coordination. Domains involved, one in βmys the other in MYBPC3, form coordinated domains (co-domains). Co-domain bilateral structure implies the possibility for a shared impact from SNV modification in either domain suggesting a correlated response to a common perturbation could identify their location. Genetic divergence over human populations is proposed to perturb SNV probability coupling that is detected by cross-correlation in 2D correlation genetics (2D-CG). SNV probability data and 2D-CG identify three critical sites, two in MYBPC3 with links to several domains across the βmys motor, and, one in βmys with links to the MYBPC3 regulatory domain. MYBPC3 sites are hinges sterically enabling regulatory interactions with βmys. The βmys site is the actin binding C-loop (residues 359-377). The C-loop is a trigger for actin-activated myosin ATPase and a contraction velocity modulator. Co-domain identification implies their spatial proximity suggesting a novel approach for in vivo protein complex structure determination.
Keywords: 2D Correlation genetics; Cardiac muscle inheritable disease; Interprotein coordinated domains; Population genetic divergence proxy; Serial founder effect; Single nucleotide variants.
© 2021. The Author(s).
Conflict of interest statement
The author has no relevant financial or non-financial interest to disclose.
Figures






Similar articles
-
Neural-symbolic hybrid model for myosin complex in cardiac ventriculum decodes structural bases for inheritable heart disease from its genetic encoding.Arch Biochem Biophys. 2025 Mar;765:110323. doi: 10.1016/j.abb.2025.110323. Epub 2025 Jan 30. Arch Biochem Biophys. 2025. PMID: 39892686
-
Cardiac and skeletal actin substrates uniquely tune cardiac myosin strain-dependent mechanics.Open Biol. 2018 Nov 21;8(11):180143. doi: 10.1098/rsob.180143. Open Biol. 2018. PMID: 30463911 Free PMC article.
-
Analytical comparison of natural and pharmaceutical ventricular myosin activators.Biochemistry. 2014 Aug 19;53(32):5298-306. doi: 10.1021/bi500730t. Epub 2014 Aug 7. Biochemistry. 2014. PMID: 25068717 Free PMC article.
-
Cardiac myosin-binding protein C (MYBPC3) in cardiac pathophysiology.Gene. 2015 Dec 1;573(2):188-97. doi: 10.1016/j.gene.2015.09.008. Epub 2015 Sep 8. Gene. 2015. PMID: 26358504 Free PMC article. Review.
-
Regulation of contraction in striated muscle.Physiol Rev. 2000 Apr;80(2):853-924. doi: 10.1152/physrev.2000.80.2.853. Physiol Rev. 2000. PMID: 10747208 Review.
Cited by
-
Eucommiae Folium and Active Compounds Protect Against Mitochondrial Dysfunction-Calcium Overload in Epileptic Hippocampal Neurons Through the Hypertrophic Cardiomyopathy Pathway.Neurochem Res. 2023 Sep;48(9):2674-2686. doi: 10.1007/s11064-023-03937-5. Epub 2023 Apr 17. Neurochem Res. 2023. PMID: 37067737
References
-
- Rayment I, Holden HM. Myosin subfragment-1: structure and function of a molecular motor. Curr Opin Struct Biol. 1993;1993:944–952. doi: 10.1016/0959-440X(93)90160-M. - DOI
LinkOut - more resources
Full Text Sources
