E3 ubiquitin ligases: styles, structures and functions
- PMID: 35006464
- PMCID: PMC8607428
- DOI: 10.1186/s43556-021-00043-2
E3 ubiquitin ligases: styles, structures and functions
Abstract
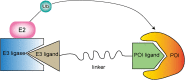
E3 ubiquitin ligases are a large family of enzymes that join in a three-enzyme ubiquitination cascade together with ubiquitin activating enzyme E1 and ubiquitin conjugating enzyme E2. E3 ubiquitin ligases play an essential role in catalyzing the ubiquitination process and transferring ubiquitin protein to attach the lysine site of targeted substrates. Importantly, ubiquitination modification is involved in almost all life activities of eukaryotes. Thus, E3 ligases might be involved in regulating various biological processes and cellular responses to stress signal associated with cancer development. Thanks to their multi-functions, E3 ligases can be a promising target of cancer therapy. A deeper understanding of the regulatory mechanisms of E3 ligases in tumorigenesis will help to find new prognostic markers and accelerate the growth of anticancer therapeutic approaches. In general, we mainly introduce the classifications of E3 ligases and their important roles in cancer progression and therapeutic functions.
Keywords: 26S proteasome degradation; Cancer progression; E3 ligases; PROTACs; Therapeutics; Ubiquitination.
© 2021. The Author(s).
Conflict of interest statement
The authors have declared that no competing interest exists.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
