Progressive loss of hepatocyte nuclear factor 4 alpha activity in chronic liver diseases in humans
- PMID: 35006629
- PMCID: PMC9762158
- DOI: 10.1002/hep.32326
Progressive loss of hepatocyte nuclear factor 4 alpha activity in chronic liver diseases in humans
Abstract
Background and aims: Hepatocyte nuclear factor 4 alpha (HNF4α) is indispensable for hepatocyte differentiation and critical for maintaining liver health. Here, we demonstrate that loss of HNF4α activity is a crucial step in the pathogenesis of chronic liver diseases (CLDs) that lead to development of HCC.
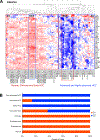
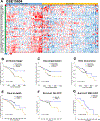
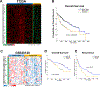
Approach and results: We developed an HNF4α target gene signature, which can accurately determine HNF4α activity, and performed an exhaustive in silico analysis using hierarchical and K-means clustering, survival, and rank-order analysis of 30 independent data sets containing over 3500 individual samples. The association of changes in HNF4α activity to CLD progression of various etiologies, including HCV- and HBV-induced liver cirrhosis (LC), NAFLD/NASH, and HCC, was determined. Results revealed a step-wise reduction in HNF4α activity with each progressive stage of pathogenesis. Cluster analysis of LC gene expression data sets using the HNF4α signature showed that loss of HNF4α activity was associated with progression of Child-Pugh class, faster decompensation, incidence of HCC, and lower survival with and without HCC. A moderate decrease in HNF4α activity was observed in NAFLD from normal liver, but a further significant decline was observed in patients from NAFLD to NASH. In HCC, loss of HNF4α activity was associated with advanced disease, increased inflammatory changes, portal vein thrombosis, and substantially lower survival.
Conclusions: In conclusion, these data indicate that loss of HNF4α function is a common event in the pathogenesis of CLDs leading to HCC and is important from both diagnostic and therapeutic standpoints.
© 2022 American Association for the Study of Liver Diseases.
Conflict of interest statement
Conflict of interest statement:
None of the authors have any conflict to declare
Figures



References
-
- Gonzalez FJ. Regulation of hepatocyte nuclear factor 4 alpha-mediated transcription. Drug Metab Pharmacokinet 2008;23:2–7. - PubMed
-
- Sladek FM. Orphan receptor HNF-4 and liver-specific gene expression. Receptor 1994;4:64. - PubMed
-
- Watt AJ, Garrison WD, Duncan SA. HNF4: a central regulator of hepatocyte differentiation and function. Hepatology 2003;37:1249–1253. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

