Protein Engineering in Ribosomally Synthesized and Post-translationally Modified Peptides (RiPPs)
- PMID: 35006671
- PMCID: PMC9454058
- DOI: 10.1021/acs.biochem.1c00714
Protein Engineering in Ribosomally Synthesized and Post-translationally Modified Peptides (RiPPs)
Abstract
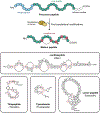
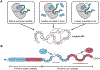
Ribosomally synthesized and post-translationally modified peptides (RiPPs) make up a rapidly growing superfamily of natural products. RiPPs exhibit an extraordinary range of structures, but they all begin as gene-encoded precursor peptides that are linear chains of amino acids produced by ribosomes. Given the gene-encoded nature of RiPP precursor peptides, the toolbox of protein engineering can be directly applied to these precursors. This Perspective will discuss examples of site-directed mutagenesis, noncanonical amino acid mutagenesis, and the construction and screening of combinatorial libraries as applied to RiPPs. These studies have led to important insights into the biosynthesis and bioactivity of RiPPs and the reengineering of RiPPs for entirely new functions.
Figures

Similar articles
-
Engineering RiPP pathways: strategies for generating complex bioactive peptides.Trends Biochem Sci. 2025 Jun;50(6):495-507. doi: 10.1016/j.tibs.2025.04.001. Epub 2025 May 6. Trends Biochem Sci. 2025. PMID: 40335383 Review.
-
Synthesis of Ribosomally Synthesized and Post-Translationally Modified Peptides Containing C-C Cross-Links.J Nat Prod. 2022 Oct 28;85(10):2519-2539. doi: 10.1021/acs.jnatprod.2c00508. Epub 2022 Sep 22. J Nat Prod. 2022. PMID: 36136399 Free PMC article. Review.
-
Expanded genetic code for the engineering of ribosomally synthetized and post-translationally modified peptide natural products (RiPPs).Curr Opin Biotechnol. 2013 Aug;24(4):591-8. doi: 10.1016/j.copbio.2013.02.026. Epub 2013 Mar 25. Curr Opin Biotechnol. 2013. PMID: 23537814 Review.
-
Ribosomally synthesized and post-translationally modified peptide natural products: new insights into the role of leader and core peptides during biosynthesis.Chemistry. 2013 Jun 10;19(24):7662-77. doi: 10.1002/chem.201300401. Epub 2013 May 10. Chemistry. 2013. PMID: 23666908 Free PMC article.
-
Genome mining for ribosomally synthesized and post-translationally modified peptides (RiPPs) in Streptomyces bacteria.J Asian Nat Prod Res. 2025 Mar;27(3):354-367. doi: 10.1080/10286020.2024.2390510. Epub 2024 Aug 14. J Asian Nat Prod Res. 2025. PMID: 39140768
Cited by
-
Mutexa: A Computational Ecosystem for Intelligent Protein Engineering.J Chem Theory Comput. 2023 Nov 14;19(21):7459-7477. doi: 10.1021/acs.jctc.3c00602. Epub 2023 Oct 12. J Chem Theory Comput. 2023. PMID: 37828731 Free PMC article. Review.
-
Large-scale Bioinformatic Study of Graspimiditides and Structural Characterization of Albusimiditide.ACS Chem Biol. 2023 Nov 17;18(11):2394-2404. doi: 10.1021/acschembio.3c00365. Epub 2023 Oct 19. ACS Chem Biol. 2023. PMID: 37856788 Free PMC article.
-
Kinetic Analysis of Cyclization by the Substrate-Tolerant Lanthipeptide Synthetase ProcM.ACS Catal. 2024 Nov 27;14(24):18310-18321. doi: 10.1021/acscatal.4c06216. eCollection 2024 Dec 20. ACS Catal. 2024. PMID: 39722886 Free PMC article.
-
Investigating the Specificity of the Dehydration and Cyclization Reactions in Engineered Lanthipeptides by Synechococcal SyncM.ACS Synth Biol. 2023 Jan 20;12(1):164-177. doi: 10.1021/acssynbio.2c00455. Epub 2022 Dec 15. ACS Synth Biol. 2023. PMID: 36520855 Free PMC article.
-
Substrate Specificity of a Methyltransferase Involved in the Biosynthesis of the Lantibiotic Cacaoidin.Biochemistry. 2024 Oct 1;63(19):2493-2505. doi: 10.1021/acs.biochem.4c00150. Epub 2024 Sep 13. Biochemistry. 2024. PMID: 39271288 Free PMC article.
References
-
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Goransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Muller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Sussmuth RD, Tagg JR, Tang G-L, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, van der Donk WA: Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Natural Product Reports 2013, 30:108–160. - PMC - PubMed
-
- Montalban-Lopez M, Scott TA, Ramesh S, Rahman IR, van Heel AJ, Viel JH, Bandarian V, Dittmann E, Genilloud O, Goto Y, Burgos MJG, Hill C, Kim S, Koehnke J, Latham JA, Link AJ, Martinez B, Nair SK, Nicolet Y, Rebuffat S, Sahl HG, Sareen D, Schmidt EW, Schmitt L, Severinov K, Sussmuth RD, Truman AW, Wang H, Weng JK, van Wezel GP, Zhang Q, Zhong J, Piel J, Mitchell DA, Kuipers OP, van der Donk WA: New developments in RiPP discovery, enzymology and engineering. Natural Product Reports 2021, 38:130–239. - PMC - PubMed
-
- Winter G, Fersht AR, Wilkinson AJ, Zoller M, Smith M: Redesigning enzyme structure by site-directed mutagenesis: tyrosyl tRNA synthetase and ATP binding. Nature 1982, 299:756–758. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

