Molecular basis of Arginine and Lysine DNA sequence-dependent thermo-stability modulation
- PMID: 35007284
- PMCID: PMC8782489
- DOI: 10.1371/journal.pcbi.1009749
Molecular basis of Arginine and Lysine DNA sequence-dependent thermo-stability modulation
Abstract
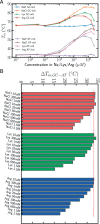
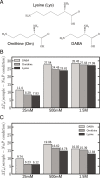
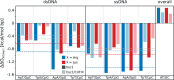
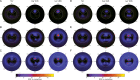
We have used a variety of theoretical and experimental techniques to study the role of four basic amino acids-Arginine, Lysine, Ornithine and L-2,4-Diaminobutyric acid-on the structure, flexibility and sequence-dependent stability of DNA. We found that the presence of organic ions stabilizes the duplexes and significantly reduces the difference in stability between AT- and GC-rich duplexes with respect to the control conditions. This suggests that these amino acids, ingredients of the primordial soup during abiogenesis, could have helped to equalize the stability of AT- and GC-rich DNA oligomers, facilitating a general non-catalysed self-replication of DNA. Experiments and simulations demonstrate that organic ions have an effect that goes beyond the general electrostatic screening, involving specific interactions along the grooves of the double helix. We conclude that organic ions, largely ignored in the DNA world, should be reconsidered as crucial structural elements far from mimics of small inorganic cations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Jorgensen WL, Pranata J. Importance of Secondary Interactions in Triply Hydrogen Bonded Complexes: Guanine-Cytosine vs Uracil-2,6-Diaminopyndine. J Am Chem Soc. 1990;112: 2008–2010. doi: 10.1021/ja00161a061 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

