Quantitative Proteomics Using Isobaric Labeling: A Practical Guide
- PMID: 35007772
- PMCID: PMC9170757
- DOI: 10.1016/j.gpb.2021.08.012
Quantitative Proteomics Using Isobaric Labeling: A Practical Guide
Abstract
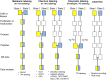
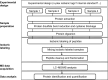
In the past decade, relative proteomic quantification using isobaric labeling technology has developed into a key tool for comparing the expression of proteins in biological samples. Although its multiplexing capacity and flexibility make this a valuable technology for addressing various biological questions, its quantitative accuracy and precision still pose significant challenges to the reliability of its quantification results. Here, we give a detailed overview of the different kinds of isobaric mass tags and the advantages and disadvantages of the isobaric labeling method. We also discuss which precautions should be taken at each step of the isobaric labeling workflow, to obtain reliable quantification results in large-scale quantitative proteomics experiments. In the last section, we discuss the broad applications of the isobaric labeling technology in biological and clinical studies, with an emphasis on thermal proteome profiling and proteogenomics.
Keywords: Isobaric labeling; Mass spectrometry; Quantitative proteomics; TMT; iTRAQ.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Bensimon A., Heck A.J., Aebersold R. Mass spectrometry-based proteomics and network biology. Annu Rev Biochem. 2012;81:379–405. - PubMed
-
- Bantscheff M., Schirle M., Sweetman G., Rick J., Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. - PubMed
-
- Bantscheff M., Lemeer S., Savitski M.M., Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404:939–965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

