A multi-dimensional evaluation of the 'NIST 1032' sample set across four forensic Y-STR multiplexes
- PMID: 35007854
- PMCID: PMC9901497
- DOI: 10.1016/j.fsigen.2021.102655
A multi-dimensional evaluation of the 'NIST 1032' sample set across four forensic Y-STR multiplexes
Abstract
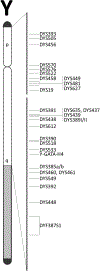
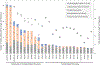
This manuscript reports Y-chromosomal short tandem repeat (Y-STR) haplotypes for 1032 male U.S. population samples across 30 Y-STR loci characterized by three capillary electrophoresis (CE) length-based kits (PowerPlex Y23 System, Yfiler Plus PCR Amplification Kit, and Investigator Argus Y-28 QS Kit) and one sequence-based kit (ForenSeq DNA Signature Prep Kit): DYF387S1, DYS19, DYS385 a/b, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, DYS439, DYS448, DYS449, DYS456, DYS458, DYS460, DYS481, DYS505, DYS518, DYS522, DYS533, DYS549, DYS570, DYS576, DYS612, DYS627, DYS635, DYS643, and Y-GATA-H4. The length-based Y-STR haplotypes include six loci that are not reported in the sequence-based kit (DYS393, DYS449, DYS456, DYS458, DYS518, and DYS627), whereas three loci included in the sequence-based kit are not present in length-based kits (DYS505, DYS522, and DYS612). For the latter, a custom multiplex was used to generate CE length-based data, allowing 1032 samples to be evaluated for concordance across the 30 Y-STR loci included in these four commercial Y-STR typing kits. Discordances between typing methods were analyzed further to assess underlying causes such as primer binding site mutations and flanking region insertions/deletions. Allele-level frequency and statistical information is provided for sequenced loci, excluding the multi-copy loci DYF387S1 and DYS385 a/b, for which locus-specific haplotype-level frequencies are provided instead. The resulting data reveals the degree of information gained through sequencing: 88% of sequenced Y-STR loci contain additional sequence-based alleles compared to length-based data, with the DYS389II locus containing the most additional alleles (51) observed by sequencing. Despite these allelic increases, only minimal improvement was observed in haplotype resolution by sequence, with all four commercial kits providing a similar ability to differentiate length-based haplotypes in this sample set. Finally, a subset of 369 male samples were compared to their corresponding additionally sequenced father samples, revealing the sequence basis for the 50 length-based changes observed, and no additional sequence-based mutations. GenBank accession numbers are reported for each unique sequence, and associated records are available in the STRSeq Y-Chromosomal STR Loci National Center for Biotechnology Information (NCBI) BioProject, accession PRJNA380347. Haplotype data is updated in the Y-STR Haplotype Reference Database (YHRD) for the 'NIST 1032' data set to now achieve the level of maximal haplotype of YHRD. All supplementary files including revisions to previously published Y-STR data are available in the NIST Public Data Repository: U.S. population data for human identification markers, DOI 10.18434/t4/1500024.
Keywords: DNA sequencing; Haplotype frequency; MPS; NGS; Short tandem repeat; Y-STR typing.
Published by Elsevier B.V.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures
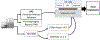
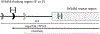

Similar articles
-
Sequence-based mutation patterns at 41 Y chromosomal STRs in 2 548 father-son pairs.Forensic Sci Res. 2023 May 31;8(2):152-162. doi: 10.1093/fsr/owad016. eCollection 2023 Jun. Forensic Sci Res. 2023. PMID: 37621447 Free PMC article.
-
Mutation analysis for 25 Y-STR markers in Japanese population.Leg Med (Tokyo). 2021 May;50:101860. doi: 10.1016/j.legalmed.2021.101860. Epub 2021 Feb 13. Leg Med (Tokyo). 2021. PMID: 33607450
-
Genetic analysis of 27 Y-chromosomal STR loci in a Zimbabwean Shona ethnic group.Leg Med (Tokyo). 2020 Mar;43:101660. doi: 10.1016/j.legalmed.2019.101660. Epub 2019 Dec 11. Leg Med (Tokyo). 2020. PMID: 31911187
-
Investigation into the sequence structure of 23 Y chromosomal STR loci using massively parallel sequencing.Forensic Sci Int Genet. 2016 Nov;25:132-141. doi: 10.1016/j.fsigen.2016.08.010. Epub 2016 Aug 28. Forensic Sci Int Genet. 2016. PMID: 27591816
-
Population genetics of 26 Y-STR loci for the Han ethnic in Hunan province, China.Int J Legal Med. 2017 Jan;131(1):115-117. doi: 10.1007/s00414-016-1411-7. Epub 2016 Jul 23. Int J Legal Med. 2017. PMID: 27448570 Review.
Cited by
-
Systematic analysis of population studies performed with the ForenSeq™ DNA Signature Prep kit.J Forensic Sci. 2025 Jul;70(4):1227-1248. doi: 10.1111/1556-4029.70057. Epub 2025 Apr 18. J Forensic Sci. 2025. PMID: 40249084 Free PMC article.
-
Development and validation of a new multiplex for upgrading Y-STRs population databases from 12 to 23 markers and its forensic casework application.Sci Rep. 2022 Dec 16;12(1):21734. doi: 10.1038/s41598-022-25785-z. Sci Rep. 2022. PMID: 36526709 Free PMC article.
-
US Population Data for 94 Identity-Informative SNP Loci.Genes (Basel). 2023 May 12;14(5):1071. doi: 10.3390/genes14051071. Genes (Basel). 2023. PMID: 37239431 Free PMC article.
-
Bed bugs, Cimex lectularius: Undercover agents in forensic investigations.J Forensic Sci. 2025 Jan;70(1):264-270. doi: 10.1111/1556-4029.15638. Epub 2024 Oct 14. J Forensic Sci. 2025. PMID: 39400324 Free PMC article.
-
Concordance study on Y-STRs typing between SeqStudio™ genetic analyzer for HID and MiSeq™ FGx forensic genomics system.Mol Biol Rep. 2023 Dec;50(12):9779-9789. doi: 10.1007/s11033-023-08808-4. Epub 2023 Oct 9. Mol Biol Rep. 2023. PMID: 37812349 Free PMC article.
References
-
- Coble MD, Hill CR, Butler JM, Haplotype data for 23 Y-chromosome markers in four U.S. population groups, Forensic Sci. Int. Genet 7 (3) (2013) e66–e68. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

