Microfluidic-Assisted Preparation of Targeted pH-Responsive Polymeric Micelles Improves Gemcitabine Effectiveness in PDAC: In Vitro Insights
- PMID: 35008170
- PMCID: PMC8750671
- DOI: 10.3390/cancers14010005
Microfluidic-Assisted Preparation of Targeted pH-Responsive Polymeric Micelles Improves Gemcitabine Effectiveness in PDAC: In Vitro Insights
Abstract
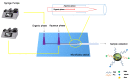
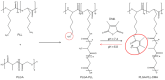
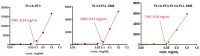
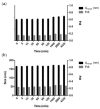
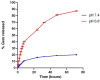
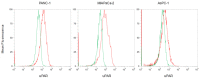
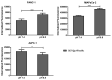
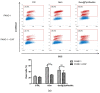
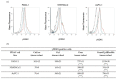
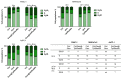
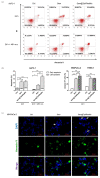
Pancreatic ductal adenocarcinoma (PDAC) represents a great challenge to the successful delivery of the anticancer drugs. The intrinsic characteristics of the PDAC microenvironment and drugs resistance make it suitable for therapeutic approaches with stimulus-responsive drug delivery systems (DDSs), such as pH, within the tumor microenvironment (TME). Moreover, the high expression of uPAR in PDAC can be exploited for a drug receptor-mediated active targeting strategy. Here, a pH-responsive and uPAR-targeted Gemcitabine (Gem) DDS, consisting of polymeric micelles (Gem@TpHResMic), was formulated by microfluidic technique to obtain a preparation characterized by a narrow size distribution, good colloidal stability, and high drug-encapsulation efficiency (EE%). The Gem@TpHResMic was able to perform a controlled Gem release in an acidic environment and to selectively target uPAR-expressing tumor cells. The Gem@TpHResMic displayed relevant cellular internalization and greater antitumor properties than free Gem in 2D and 3D models of pancreatic cancer, by generating massive damage to DNA, in terms of H2AX phosphorylation and apoptosis induction. Further investigation into the physiological model of PDAC, obtained by a co-culture of tumor spheroids and cancer-associated fibroblast (CAF), highlighted that the micellar system enhanced the antitumor potential of Gem, and was demonstrated to overcome the TME-dependent drug resistance. In vivo investigation is warranted to consider this new DDS as a new approach to overcome drug resistance in PDAC.
Keywords: active drug targeting; controlled release; drug delivery; drug resistance; pH-responsiveness; pancreatic ductal adenocarcinoma; tumor microenvironment; uPAR.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- AIOM. [(accessed on 15 September 2021)]. Available online: https://www.aiom.it/wp-content/uploads/2020/10/2020_LG_AIOM_Pancreas.pdf.
-
- Porcelli L., Iacobazzi R.M., Di Fonte R., Serratì S., Intini A., Solimando A.G., Brunetti O., Calabrese A., Leonetti F., Azzariti A., et al. CAFs and TGF-β Signaling Activation by Mast Cells Contribute to Resistance to Gemcitabine/Nabpaclitaxel in Pancreatic Cancer. Cancers. 2019;11:330. doi: 10.3390/cancers11030330. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

