Manganese Is a Strong Specific Activator of the RNA Synthetic Activity of Human Polη
- PMID: 35008656
- PMCID: PMC8745064
- DOI: 10.3390/ijms23010230
Manganese Is a Strong Specific Activator of the RNA Synthetic Activity of Human Polη
Abstract
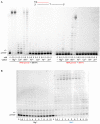
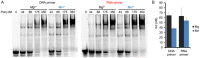
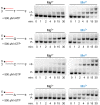
DNA polymerase η (Polη) is a translesion synthesis polymerase that can bypass different DNA lesions with varying efficiency and fidelity. Its most well-known function is the error-free bypass of ultraviolet light-induced cyclobutane pyrimidine dimers. The lack of this unique ability in humans leads to the development of a cancer-predisposing disease, the variant form of xeroderma pigmentosum. Human Polη can insert rNTPs during DNA synthesis, though with much lower efficiency than dNTPs, and it can even extend an RNA chain with ribonucleotides. We have previously shown that Mn2+ is a specific activator of the RNA synthetic activity of yeast Polη that increases the efficiency of the reaction by several thousand-fold over Mg2+. In this study, our goal was to investigate the metal cofactor dependence of RNA synthesis by human Polη. We found that out of the investigated metal cations, only Mn2+ supported robust RNA synthesis. Steady state kinetic analysis showed that Mn2+ activated the reaction a thousand-fold compared to Mg2+, even during DNA damage bypass opposite 8-oxoG and TT dimer. Our results revealed a two order of magnitude higher affinity of human Polη towards ribonucleotides in the presence of Mn2+ compared to Mg2+. It is noteworthy that activation occurred without lowering the base selectivity of the enzyme on undamaged templates, whereas the fidelity decreased across a TT dimer. In summary, our data strongly suggest that, like with its yeast homolog, Mn2+ is the proper metal cofactor of hPolη during RNA chain extension, and selective metal cofactor utilization contributes to switching between its DNA and RNA synthetic activities.
Keywords: RNA extension; enzyme kinetics; human polymerase η; manganese; translesion synthesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Pan Q., Wang L., Liu Y., Li M., Zhang Y., Peng W., Deng T., Peng M.-L., Jiang J.-Q., Tang J., et al. Knockdown of POLQ interferes the development and progression of hepatocellular carcinoma through regulating cell proliferation, apoptosis and migration. Cancer Cell Int. 2021;21:482. doi: 10.1186/s12935-021-02178-2. - DOI - PMC - PubMed
-
- Ohkumo T., Kondo Y., Yokoi M., Tsukamoto T., Yamada A., Sugimoto T., Kanao R., Higashi Y., Kondoh H., Tatematsu M., et al. UV-B Radiation Induces Epithelial Tumors in Mice Lacking DNA Polymerase η and Mesenchymal Tumors in Mice Deficient for DNA Polymerase ι. Mol. Cell. Biol. 2006;26:7696–7706. doi: 10.1128/MCB.01076-06. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

