Dietary Interventions Ameliorate Infectious Colitis by Restoring the Microbiome and Promoting Stem Cell Proliferation in Mice
- PMID: 35008767
- PMCID: PMC8745185
- DOI: 10.3390/ijms23010339
Dietary Interventions Ameliorate Infectious Colitis by Restoring the Microbiome and Promoting Stem Cell Proliferation in Mice
Abstract
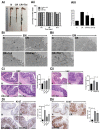
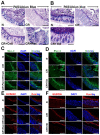
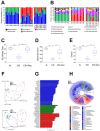
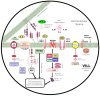
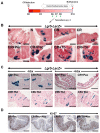
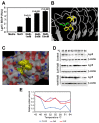
Decreases in short-chain-fatty-acids (SCFAs) are linked to inflammatory bowel disease (IBD). Yet, the mechanisms through which SCFAs promote wound healing, orchestrated by intestinal stem cells, are poorly understood. We discovered that, in mice with Citrobacter rodentium (CR)-induced infectious colitis, treatment with Pectin and Tributyrin diets reduced the severity of colitis by restoring Firmicutes and Bacteroidetes and by increasing mucus production. RNA-seq in young adult mouse colon (YAMC) cells identified higher expression of Lgr4, Lgr6, DCLK1, Muc2, and SIGGIR after Butyrate treatment. Lineage tracing in CR-infected Lgr5-EGFP-IRES-CreERT2/ROSA26-LacZ (Lgr5-R) mice also revealed an expansion of LacZ-labeled Lgr5(+) stem cells in the colons of both Pectin and Tributyrin-treated mice compared to control. Interestingly, gut microbiota was required for Pectin but not Tributyrin-induced Lgr5(+) stem cell expansion. YAMC cells treated with sodium butyrate exhibited increased Lgr5 promoter reporter activity due to direct Butyrate binding with Lgr5 at -4.0 Kcal/mol, leading to thermal stabilization. Upon ChIP-seq, H3K4me3 increased near Lgr5 transcription start site that contained the consensus binding motif for a transcriptional activator of Lgr5 (SPIB). Thus, a multitude of effects on gut microbiome, differential gene expression, and/or expansion of Lgr5(+) stem cells seem to underlie amelioration of colitis following dietary intervention.
Keywords: Citrobacter rodentium; Pectin; Tributyrin; bacterial infection; colitis; dietary intervention; gut microbiome; intestinal stem cells; lgr5; short-chain-fatty-acids (SCFAs).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Candela M., Perna F., Carnevali P., Vitali B., Ciati R., Gionchetti P., Rizzello F., Campieri M., Brigidi P. Interaction of probiotic Lactobacillus and Bifidobacterium strains with human intestinal epithelial cells: Adhesion properties, competition against enteropathogens and modulation of IL-8 production. Int. J. Food Microbiol. 2008;125:286–292. doi: 10.1016/j.ijfoodmicro.2008.04.012. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

