Vypal2: A Versatile Peptide Ligase for Precision Tailoring of Proteins
- PMID: 35008882
- PMCID: PMC8745061
- DOI: 10.3390/ijms23010458
Vypal2: A Versatile Peptide Ligase for Precision Tailoring of Proteins
Abstract
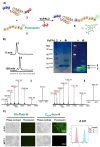
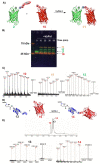
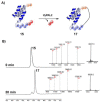
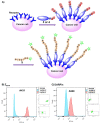
The last two decades have seen an increasing demand for new protein-modification methods from the biotech industry and biomedical research communities. Owing to their mild aqueous reaction conditions, enzymatic methods based on the use of peptide ligases are particularly desirable. In this regard, the recently discovered peptidyl Asx-specific ligases (PALs) have emerged as powerful biotechnological tools in recent years. However, as a new class of peptide ligases, their scope and application remain underexplored. Herein, we report the use of a new PAL, VyPAL2, for a diverse range of protein modifications. We successfully showed that VyPAL2 was an efficient biocatalyst for protein labelling, inter-protein ligation, and protein cyclization. The labelled or cyclized protein ligands remained functionally active in binding to their target receptors. We also demonstrated on-cell labelling of protein ligands pre-bound to cellular receptors and cell-surface engineering via modifying a covalently anchored peptide substrate pre-installed on cell-surface glycans. Together, these examples firmly establish Asx-specific ligases, such as VyPAL2, as the biocatalysts of the future for site-specific protein modification, with a myriad of applications in basic research and drug discovery.
Keywords: VyPAL2; biocatalysts; cell surface labeling; protein cyclization; protein labeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






Similar articles
-
PAL-Mediated Ligation for Protein and Cell-Surface Modification.Methods Mol Biol. 2022;2530:177-193. doi: 10.1007/978-1-0716-2489-0_13. Methods Mol Biol. 2022. PMID: 35761050
-
Engineering protein theranostics using bio-orthogonal asparaginyl peptide ligases.Theranostics. 2021 Apr 3;11(12):5863-5875. doi: 10.7150/thno.53615. eCollection 2021. Theranostics. 2021. PMID: 33897886 Free PMC article.
-
Structural basis for proenzyme maturation, substrate recognition, and ligation by a hyperactive peptide asparaginyl ligase.Plant Cell. 2022 Nov 29;34(12):4936-4949. doi: 10.1093/plcell/koac281. Plant Cell. 2022. PMID: 36099055 Free PMC article.
-
Make it or break it: Plant AEPs on stage in biotechnology.Biotechnol Adv. 2020 Dec;45:107651. doi: 10.1016/j.biotechadv.2020.107651. Epub 2020 Oct 23. Biotechnol Adv. 2020. PMID: 33141031 Review.
-
Challenges in the use of sortase and other peptide ligases for site-specific protein modification.Chem Soc Rev. 2022 May 23;51(10):4121-4145. doi: 10.1039/d0cs01148g. Chem Soc Rev. 2022. PMID: 35510539 Free PMC article. Review.
Cited by
-
An Analysis Regarding the Association Between DAZ Interacting Zinc Finger Protein 1 (DZIP1) and Colorectal Cancer (CRC).Mol Biotechnol. 2025 Feb;67(2):527-547. doi: 10.1007/s12033-024-01065-1. Epub 2024 Feb 9. Mol Biotechnol. 2025. PMID: 38334905
-
Chronic disease management via modulation of cellular signaling by phytoestrogen Bavachin.Mol Biol Rep. 2024 Aug 19;51(1):921. doi: 10.1007/s11033-024-09849-z. Mol Biol Rep. 2024. PMID: 39158613 Review.
-
Increased protein expression of interleukin-10 and its signalling molecules in colon cancer progression: potential prognostic and therapeutic targets.Discov Oncol. 2025 Apr 29;16(1):637. doi: 10.1007/s12672-025-02452-z. Discov Oncol. 2025. PMID: 40299210 Free PMC article.
-
Climate change and plant genomic plasticity.Theor Appl Genet. 2025 Aug 27;138(9):231. doi: 10.1007/s00122-025-05010-x. Theor Appl Genet. 2025. PMID: 40864264 Free PMC article. Review.
-
Drought stress memory in maize: understanding and harnessing the past for future resilience.Plant Cell Rep. 2025 Apr 25;44(5):101. doi: 10.1007/s00299-025-03494-x. Plant Cell Rep. 2025. PMID: 40278890 Review.
References
-
- Xu S., Zhao Z., Zhao J. Recent advances in enzyme-mediated peptide ligation. Chin. Chem. Lett. 2018;29:1009–1016. doi: 10.1016/j.cclet.2018.05.024. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

