Pyranose Ring Puckering Thermodynamics for Glycan Monosaccharides Associated with Vertebrate Proteins
- PMID: 35008898
- PMCID: PMC8745717
- DOI: 10.3390/ijms23010473
Pyranose Ring Puckering Thermodynamics for Glycan Monosaccharides Associated with Vertebrate Proteins
Abstract
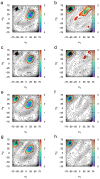
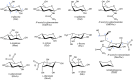
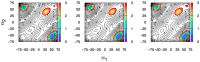
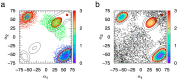
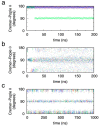
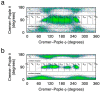
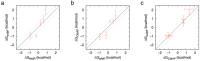
The conformational properties of carbohydrates can contribute to protein structure directly through covalent conjugation in the cases of glycoproteins and proteoglycans and indirectly in the case of transmembrane proteins embedded in glycolipid-containing bilayers. However, there continue to be significant challenges associated with experimental structural biology of such carbohydrate-containing systems. All-atom explicit-solvent molecular dynamics simulations provide a direct atomic resolution view of biomolecular dynamics and thermodynamics, but the accuracy of the results depends on the quality of the force field parametrization used in the simulations. A key determinant of the conformational properties of carbohydrates is ring puckering. Here, we applied extended system adaptive biasing force (eABF) all-atom explicit-solvent molecular dynamics simulations to characterize the ring puckering thermodynamics of the ten common pyranose monosaccharides found in vertebrate biology (as represented by the CHARMM carbohydrate force field). The results, along with those for idose, demonstrate that the CHARMM force field reliably models ring puckering across this diverse set of molecules, including accurately capturing the subtle balance between 4C1 and 1C4 chair conformations in the cases of iduronate and of idose. This suggests the broad applicability of the force field for accurate modeling of carbohydrate-containing vertebrate biomolecules such as glycoproteins, proteoglycans, and glycolipids.
Keywords: GalNAc; GlcNAc; Neu5Ac; fucose; galactose; glucose; glucuronate; iduronate; mannose; tetrahydropyran; xylose.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures
Similar articles
-
On the Use of PDB X-Ray Crystal Structures as Force Field Target and Validation Data for Pyranose Ring Puckering.J Comput Chem. 2025 Apr 30;46(11):e70110. doi: 10.1002/jcc.70110. J Comput Chem. 2025. PMID: 40251812
-
Impact of Polarization on the Ring Puckering Dynamics of Hexose Monosaccharides.J Chem Inf Model. 2023 Jan 9;63(1):208-223. doi: 10.1021/acs.jcim.2c01286. Epub 2022 Dec 7. J Chem Inf Model. 2023. PMID: 36475659
-
Additive empirical force field for hexopyranose monosaccharides.J Comput Chem. 2008 Nov 30;29(15):2543-64. doi: 10.1002/jcc.21004. J Comput Chem. 2008. PMID: 18470966 Free PMC article.
-
A perspective on the primary and three-dimensional structures of carbohydrates.Carbohydr Res. 2013 Aug 30;378:123-32. doi: 10.1016/j.carres.2013.02.005. Epub 2013 Feb 24. Carbohydr Res. 2013. PMID: 23522728 Review.
-
Bioinformatics and molecular modeling in glycobiology.Cell Mol Life Sci. 2010 Aug;67(16):2749-72. doi: 10.1007/s00018-010-0352-4. Epub 2010 Apr 4. Cell Mol Life Sci. 2010. PMID: 20364395 Free PMC article. Review.
Cited by
-
Generating 3D Models of Carbohydrates with GLYCAM-Web.bioRxiv [Preprint]. 2025 May 9:2025.05.08.652828. doi: 10.1101/2025.05.08.652828. bioRxiv. 2025. PMID: 40654779 Free PMC article. Preprint.
-
Atomic-Resolution Experimental Structural Biology and Molecular Dynamics Simulations of Hyaluronan and Its Complexes.Molecules. 2022 Oct 26;27(21):7276. doi: 10.3390/molecules27217276. Molecules. 2022. PMID: 36364098 Free PMC article. Review.
-
Thrombin-derived C-terminal peptides bind and form aggregates with sulfated glycosaminoglycans.Heliyon. 2024 Aug 2;10(16):e35703. doi: 10.1016/j.heliyon.2024.e35703. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39229523 Free PMC article.
-
Refinement of the Drude Polarizable Force Field for Hexose Monosaccharides: Capturing Ring Conformational Dynamics with Enhanced Accuracy.J Chem Theory Comput. 2024 Oct 22;20(20):9161-9177. doi: 10.1021/acs.jctc.4c00656. Epub 2024 Oct 9. J Chem Theory Comput. 2024. PMID: 39383338
-
Assortment of Frontiers in Protein Science.Int J Mol Sci. 2022 Mar 28;23(7):3685. doi: 10.3390/ijms23073685. Int J Mol Sci. 2022. PMID: 35409045 Free PMC article.
References
-
- Schnaar R.L., Kinoshita T. Glycosphingolipids. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., et al., editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2015. pp. 125–135. - DOI - PubMed
-
- Seeberger P.H. Monosaccharide Diversity. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., et al., editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2017. pp. 19–30. - DOI
-
- Stanley P., Taniguchi N., Aebi M. N-Glycans. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., et al., editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2015. pp. 99–111. - DOI - PubMed
-
- Brockhausen I., Stanley P. O-GalNAc Glycans. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., et al., editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2015. pp. 113–123. - DOI - PubMed
-
- Lindahl U., Couchman J., Kimata K., Esko J.D. Proteoglycans and Sulfated Glycosaminoglycans. In: Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., et al., editors. Essentials of Glycobiology. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2015. pp. 207–221. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

