Regulation of Heterogenous LexA Expression in Staphylococcus aureus by an Antisense RNA Originating from Transcriptional Read-Through upon Natural Mispairings in the sbrB Intrinsic Terminator
- PMID: 35009002
- PMCID: PMC8745188
- DOI: 10.3390/ijms23010576
Regulation of Heterogenous LexA Expression in Staphylococcus aureus by an Antisense RNA Originating from Transcriptional Read-Through upon Natural Mispairings in the sbrB Intrinsic Terminator
Abstract
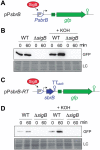
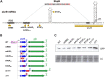
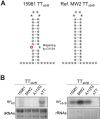
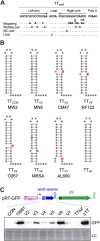
Bacterial genomes are pervasively transcribed, generating a wide variety of antisense RNAs (asRNAs). Many of them originate from transcriptional read-through events (TREs) during the transcription termination process. Previous transcriptome analyses revealed that the lexA gene from Staphylococcus aureus, which encodes the main SOS response regulator, is affected by the presence of an asRNA. Here, we show that the lexA antisense RNA (lexA-asRNA) is generated by a TRE on the intrinsic terminator (TTsbrB) of the sbrB gene, which is located downstream of lexA, in the opposite strand. Transcriptional read-through occurs by a natural mutation that destabilizes the TTsbrB structure and modifies the efficiency of the intrinsic terminator. Restoring the mispairing mutation in the hairpin of TTsbrB prevented lexA-asRNA transcription. The level of lexA-asRNA directly correlated with cellular stress since the expressions of sbrB and lexA-asRNA depend on the stress transcription factor SigB. Comparative analyses revealed strain-specific nucleotide polymorphisms within TTsbrB, suggesting that this TT could be prone to accumulating natural mutations. A genome-wide analysis of TREs suggested that mispairings in TT hairpins might provide wider transcriptional connections with downstream genes and, ultimately, transcriptomic variability among S. aureus strains.
Keywords: Staphylococcus aureus; antisense RNA; lexA; post-transcriptional regulation; transcriptional read-through; transcriptional termination.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses or interpretation of data, in the writing of the manuscript or in the decision to publish the results.
Figures


Similar articles
-
Antisense transcription is pervasive but rarely conserved in enteric bacteria.mBio. 2012 Aug 7;3(4):e00156-12. doi: 10.1128/mBio.00156-12. Print 2012. mBio. 2012. PMID: 22872780 Free PMC article.
-
A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide.Nat Struct Mol Biol. 2011 Dec 25;19(1):105-12. doi: 10.1038/nsmb.2193. Nat Struct Mol Biol. 2011. PMID: 22198463
-
Clp-dependent proteolysis of the LexA N-terminal domain in Staphylococcus aureus.Microbiology (Reading). 2011 Mar;157(Pt 3):677-684. doi: 10.1099/mic.0.043794-0. Epub 2010 Dec 23. Microbiology (Reading). 2011. PMID: 21183573
-
cis-antisense RNA, another level of gene regulation in bacteria.Microbiol Mol Biol Rev. 2011 Jun;75(2):286-300. doi: 10.1128/MMBR.00032-10. Microbiol Mol Biol Rev. 2011. PMID: 21646430 Free PMC article. Review.
-
Post-transcriptional inducible gene regulation by natural antisense RNA.Front Biosci (Landmark Ed). 2015 Jan 1;20(1):1-36. doi: 10.2741/4297. Front Biosci (Landmark Ed). 2015. PMID: 25553439 Review.
Cited by
-
BacTermFinder: a comprehensive and general bacterial terminator finder using a CNN ensemble.NAR Genom Bioinform. 2025 Mar 8;7(1):lqaf016. doi: 10.1093/nargab/lqaf016. eCollection 2025 Mar. NAR Genom Bioinform. 2025. PMID: 40060369 Free PMC article.
References
-
- Lasa I., Toledo-Arana A., Dobin A., Villanueva M., de los Mozos I.R., Vergara-Irigaray M., Segura V., Fagegaltier D., Penadés J.R., Valle J., et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc. Natl. Acad. Sci. USA. 2011;108:20172–20177. doi: 10.1073/pnas.1113521108. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

