Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation
- PMID: 35009056
- PMCID: PMC8747350
- DOI: 10.3390/plants11010051
Hairy CRISPR: Genome Editing in Plants Using Hairy Root Transformation
Abstract
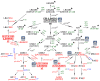
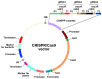
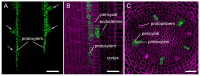
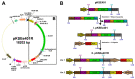
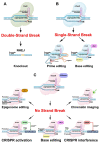
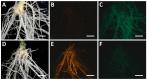
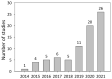
CRISPR/Cas-mediated genome editing is a powerful tool of plant functional genomics. Hairy root transformation is a rapid and convenient approach for obtaining transgenic roots. When combined, these techniques represent a fast and effective means of studying gene function. In this review, we outline the current state of the art reached by the combination of these approaches over seven years. Additionally, we discuss the origins of different Agrobacterium rhizogenes strains that are widely used for hairy root transformation; the components of CRISPR/Cas vectors, such as the promoters that drive Cas or gRNA expression, the types of Cas nuclease, and selectable and screenable markers; and the application of CRISPR/Cas genome editing in hairy roots. The modification of the already known vector pKSE401 with the addition of the rice translational enhancer OsMac3 and the gene encoding the fluorescent protein DsRed1 is also described.
Keywords: Agrobacterium strains; CRISPR/Cas; OsMac3; genome editing; hairy root transformation; pKSEe401R; vector construction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Ribozyme-mediated CRISPR/Cas9 gene editing in pyrethrum (Tanacetum cinerariifolium) hairy roots using a RNA polymerase II-dependent promoter.Plant Methods. 2022 Mar 16;18(1):32. doi: 10.1186/s13007-022-00863-5. Plant Methods. 2022. PMID: 35292048 Free PMC article.
-
Combination of Hairy Root and Whole-Plant Transformation Protocols to Achieve Efficient CRISPR/Cas9 Genome Editing in Soybean.Plants (Basel). 2023 Feb 23;12(5):1017. doi: 10.3390/plants12051017. Plants (Basel). 2023. PMID: 36903878 Free PMC article.
-
CRISPR/Cas9 in Planta Hairy Root Transformation: A Powerful Platform for Functional Analysis of Root Traits in Soybean.Plants (Basel). 2022 Apr 12;11(8):1044. doi: 10.3390/plants11081044. Plants (Basel). 2022. PMID: 35448772 Free PMC article. Review.
-
Highly efficient Agrobacterium rhizogenes-mediated hairy root transformation for gene functional and gene editing analysis in soybean.Plant Methods. 2021 Jul 10;17(1):73. doi: 10.1186/s13007-021-00778-7. Plant Methods. 2021. PMID: 34246291 Free PMC article.
-
Agrobacterium-mediated delivery of CRISPR/Cas reagents for genome editing in plants enters an era of ternary vector systems.Sci China Life Sci. 2020 Oct;63(10):1491-1498. doi: 10.1007/s11427-020-1685-9. Epub 2020 Mar 27. Sci China Life Sci. 2020. PMID: 32279281 Review.
Cited by
-
Callus Induction Followed by Regeneration and Hairy Root Induction in Common Buckwheat.Methods Mol Biol. 2024;2791:1-14. doi: 10.1007/978-1-0716-3794-4_1. Methods Mol Biol. 2024. PMID: 38532087
-
Clustered regularly interspaced short palindromic repeats/CRISPR-associated protein and hairy roots: a perfect match for gene functional analysis and crop improvement.Curr Opin Biotechnol. 2023 Feb;79:102876. doi: 10.1016/j.copbio.2022.102876. Epub 2023 Jan 6. Curr Opin Biotechnol. 2023. PMID: 36621223 Free PMC article. Review.
-
Applying conventional and cell-type-specific CRISPR/Cas9 genome editing in legume plants.aBIOTECH. 2024 Dec 16;6(2):346-360. doi: 10.1007/s42994-024-00190-4. eCollection 2025 Jun. aBIOTECH. 2024. PMID: 40641645 Free PMC article. Review.
-
Injection-based hairy root induction and plant regeneration techniques in Brassicaceae.Plant Methods. 2024 Feb 17;20(1):29. doi: 10.1186/s13007-024-01150-1. Plant Methods. 2024. PMID: 38368430 Free PMC article.
-
Agrobacterium rhizogenes-Mediated Hairy Root Genetic Transformation Using Agrobacterium Gel Inoculation and RUBY Reporter Enables Efficient Gene Function Analysis in Sacha Inchi (Plukenetia volubilis).Int J Mol Sci. 2025 Mar 11;26(6):2496. doi: 10.3390/ijms26062496. Int J Mol Sci. 2025. PMID: 40141141 Free PMC article.
References
-
- Somssich M. A short history of plant transformation. Peer J. 2019;7:1–28. doi: 10.7287/peerj.preprints.27556v2. - DOI
-
- Young J.M., Kuykendall L.D., Martínez-Romero E., Kerr A., Sawada H. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola and R. vitis. Int. J. Syst. Evol. Microbiol. 2001;51:89–103. doi: 10.1099/00207713-51-1-89. - DOI - PubMed
-
- Porter J.R., Flores H. Host range and implications of plant infection by Agrobacterium rhizogenes. Crit. Rev. Plant. Sci. 1991;10:387–421. doi: 10.1080/07352689109382318. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

