Regulation of the Later Stages of Nodulation Stimulated by IPD3/CYCLOPS Transcription Factor and Cytokinin in Pea Pisum sativum L
- PMID: 35009060
- PMCID: PMC8747635
- DOI: 10.3390/plants11010056
Regulation of the Later Stages of Nodulation Stimulated by IPD3/CYCLOPS Transcription Factor and Cytokinin in Pea Pisum sativum L
Abstract
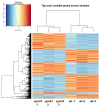
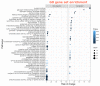
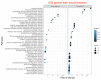
The IPD3/CYCLOPS transcription factor was shown to be involved in the regulation of nodule primordia development and subsequent stages of nodule differentiation. In contrast to early stages, the stages related to nodule differentiation remain less studied. Recently, we have shown that the accumulation of cytokinin at later stages may significantly impact nodule development. This conclusion was based on a comparative analysis of cytokinin localization between pea wild type and ipd3/cyclops mutants. However, the role of cytokinin at these later stages of nodulation is still far from understood. To determine a set of genes involved in the regulation of later stages of nodule development connected with infection progress, intracellular accommodation, as well as plant tissue and bacteroid differentiation, the RNA-seq analysis of pea mutant SGEFix--2 (sym33) nodules impaired in these processes compared to wild type SGE nodules was performed. To verify cytokinin's influence on late nodule development stages, the comparative RNA-seq analysis of SGEFix--2 (sym33) mutant plants treated with cytokinin was also conducted. Findings suggest a significant role of cytokinin in the regulation of later stages of nodule development.
Keywords: IPD3/CYCLOPS transcription factor; Pisum sativum; cytokinin; gene expression; transcriptomic data.
Conflict of interest statement
The authors declare they have no competing interests.
Figures





Similar articles
-
Exogenously Applied Cytokinin Altered the Bacterial Release and Subsequent Stages of Nodule Development in Pea Ipd3/Cyclops Mutant.Plants (Basel). 2023 Feb 2;12(3):657. doi: 10.3390/plants12030657. Plants (Basel). 2023. PMID: 36771742 Free PMC article.
-
Mutational analysis indicates that abnormalities in rhizobial infection and subsequent plant cell and bacteroid differentiation in pea (Pisum sativum) nodules coincide with abnormal cytokinin responses and localization.Ann Bot. 2020 May 13;125(6):905-923. doi: 10.1093/aob/mcaa022. Ann Bot. 2020. PMID: 32198503 Free PMC article.
-
The pea (Pisum sativum L.) genes sym33 and sym40 control infection thread formation and root nodule function.Mol Gen Genet. 1998 Sep;259(5):491-503. doi: 10.1007/s004380050840. Mol Gen Genet. 1998. PMID: 9790580
-
Symbiotic Regulatory Genes Controlling Nodule Development in Pisum sativum L.Plants (Basel). 2020 Dec 9;9(12):1741. doi: 10.3390/plants9121741. Plants (Basel). 2020. PMID: 33317178 Free PMC article. Review.
-
[Comparative genetics and evolutionary morphology of symbiosis formed by plants with nitrogen-fixing microbes and endomycorrhizal fungi].Zh Obshch Biol. 2002 Nov-Dec;63(6):451-72. Zh Obshch Biol. 2002. PMID: 12510586 Review. Russian.
Cited by
-
Exogenously Applied Cytokinin Altered the Bacterial Release and Subsequent Stages of Nodule Development in Pea Ipd3/Cyclops Mutant.Plants (Basel). 2023 Feb 2;12(3):657. doi: 10.3390/plants12030657. Plants (Basel). 2023. PMID: 36771742 Free PMC article.
-
Research Progress on Gene Regulation of Plant Floral Organogenesis.Genes (Basel). 2025 Jan 12;16(1):79. doi: 10.3390/genes16010079. Genes (Basel). 2025. PMID: 39858626 Free PMC article. Review.
-
GmWRKY33a is a hub gene responsive to brassinosteroid signaling that suppresses nodulation in soybean (Glycine max).Front Plant Sci. 2025 Jan 16;15:1507307. doi: 10.3389/fpls.2024.1507307. eCollection 2024. Front Plant Sci. 2025. PMID: 39886690 Free PMC article.
-
Genes Associated with Biological Nitrogen Fixation Efficiency Identified Using RNA Sequencing in Red Clover (Trifolium pratense L.).Life (Basel). 2022 Nov 25;12(12):1975. doi: 10.3390/life12121975. Life (Basel). 2022. PMID: 36556339 Free PMC article.
References
-
- Radutoiu S., Madsen L.H., Madsen E.B., Jurkiewicz A., Fukai E., Quistgaard E.M.H., Albrektsen A.S., James E.K., Thirup S., Stougaard J. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26:3923–3935. doi: 10.1038/sj.emboj.7601826. - DOI - PMC - PubMed
-
- Broghammer A., Krusell L., Blaise M., Sauer J., Sullivan J.T., Maolanon N., Vinther M., Lorentzen A., Madsen E.B., Jensen K.J., et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. - DOI - PMC - PubMed
-
- Amor B.B., Shaw S.L., Oldroyd G.E.D., Maillet F., Penmetsa R.V., Cook D., Long S.R., Denarie J., Gough C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003;34:495–506. doi: 10.1046/j.1365-313X.2003.01743.x. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

