The Omics Hunt for Novel Molecular Markers of Resistance to Phytophthora infestans
- PMID: 35009065
- PMCID: PMC8747139
- DOI: 10.3390/plants11010061
The Omics Hunt for Novel Molecular Markers of Resistance to Phytophthora infestans
Abstract
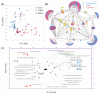
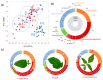
Wild Solanum accessions are a treasured source of resistance against pathogens, including oomycete Phytophthora infestans, causing late blight disease. Here, Solanum pinnatisectum, Solanum tuberosum, and the somatic hybrid between these two lines were analyzed, representing resistant, susceptible, and moderately resistant genotypes, respectively. Proteome and metabolome analyses showed that the infection had the highest impact on leaves of the resistant plant and indicated, among others, an extensive remodeling of the leaf lipidome. The lipidome profiling confirmed an accumulation of glycerolipids, a depletion in the total pool of glycerophospholipids, and showed considerable differences between the lipidome composition of resistant and susceptible genotypes. The analysis of putative resistance markers pinpointed more than 100 molecules that positively correlated with resistance including phenolics and cysteamine, a compound with known antimicrobial activity. Putative resistance protein markers were targeted in an additional 12 genotypes with contrasting resistance to P. infestans. At least 27 proteins showed a negative correlation with the susceptibility including HSP70-2, endochitinase B, WPP domain-containing protein, and cyclase 3. In summary, these findings provide insights into molecular mechanisms of resistance against P. infestans and present novel targets for selective breeding.
Keywords: Oomycetes; Oomycota; Phytophthora infestans; late blight; lipidome; metabolome; proteome; resistance; resistance markers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

