DeepLRR: An Online Webserver for Leucine-Rich-Repeat Containing Protein Characterization Based on Deep Learning
- PMID: 35009139
- PMCID: PMC8796025
- DOI: 10.3390/plants11010136
DeepLRR: An Online Webserver for Leucine-Rich-Repeat Containing Protein Characterization Based on Deep Learning
Abstract
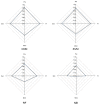
Members of the leucine-rich repeat (LRR) superfamily play critical roles in multiple biological processes. As the LRR unit sequence is highly variable, accurately predicting the number and location of LRR units in proteins is a highly challenging task in the field of bioinformatics. Existing methods still need to be improved, especially when it comes to similarity-based methods. We introduce our DeepLRR method based on a convolutional neural network (CNN) model and LRR features to predict the number and location of LRR units in proteins. We compared DeepLRR with six existing methods using a dataset containing 572 LRR proteins and it outperformed all of them when it comes to overall F1 score. In addition, DeepLRR has integrated identifying plant disease-resistance proteins (NLR, LRR-RLK, LRR-RLP) and non-canonical domains. With DeepLRR, 223, 191 and 183 LRR-RLK genes in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa ssp. Japonica) and tomato (Solanum lycopersicum) genomes were re-annotated, respectively. Chromosome mapping and gene cluster analysis revealed that 24.2% (54/223), 29.8% (57/191) and 16.9% (31/183) of LRR-RLK genes formed gene cluster structures in Arabidopsis, rice and tomato, respectively. Finally, we explored the evolutionary relationship and domain composition of LRR-RLK genes in each plant and distributions of known receptor and co-receptor pairs. This provides a new perspective for the identification of potential receptors and co-receptors.
Keywords: LRR domain; deep learning; plant disease-resistance genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Comparative analysis of evolutionary dynamics of genes encoding leucine-rich repeat receptor-like kinase between rice and Arabidopsis.Genetica. 2011 Aug;139(8):1023-32. doi: 10.1007/s10709-011-9604-y. Epub 2011 Aug 31. Genetica. 2011. PMID: 21879323
-
Molecular dissection of the response of a rice leucine-rich repeat receptor-like kinase (LRR-RLK) gene to abiotic stresses.J Plant Physiol. 2014 Nov 1;171(17):1645-53. doi: 10.1016/j.jplph.2014.08.002. Epub 2014 Aug 13. J Plant Physiol. 2014. PMID: 25173451
-
Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants.BMC Evol Biol. 2017 Feb 7;17(1):47. doi: 10.1186/s12862-017-0891-5. BMC Evol Biol. 2017. PMID: 28173747 Free PMC article.
-
Receptor-like Kinases (LRR-RLKs) in Response of Plants to Biotic and Abiotic Stresses.Plants (Basel). 2022 Oct 10;11(19):2660. doi: 10.3390/plants11192660. Plants (Basel). 2022. PMID: 36235526 Free PMC article. Review.
-
Plant receptor-like serine threonine kinases: roles in signaling and plant defense.Mol Plant Microbe Interact. 2008 May;21(5):507-17. doi: 10.1094/MPMI-21-5-0507. Mol Plant Microbe Interact. 2008. PMID: 18393610 Review.
Cited by
-
Diversity and features of proteins with structural repeats.Biophys Rev. 2023 Sep 7;15(5):1159-1169. doi: 10.1007/s12551-023-01130-0. eCollection 2023 Oct. Biophys Rev. 2023. PMID: 37974986 Free PMC article. Review.
-
Unmasking the invaders: NLR-mal function in plant defense.Front Plant Sci. 2023 Nov 20;14:1307294. doi: 10.3389/fpls.2023.1307294. eCollection 2023. Front Plant Sci. 2023. PMID: 38078096 Free PMC article. Review.
-
Network analyses predict major regulators of resistance to early blight disease complex in tomato.BMC Plant Biol. 2024 Jul 6;24(1):641. doi: 10.1186/s12870-024-05366-0. BMC Plant Biol. 2024. PMID: 38971719 Free PMC article.
-
NLRexpress-A bundle of machine learning motif predictors-Reveals motif stability underlying plant Nod-like receptors diversity.Front Plant Sci. 2022 Sep 15;13:975888. doi: 10.3389/fpls.2022.975888. eCollection 2022. Front Plant Sci. 2022. PMID: 36186050 Free PMC article.
-
Genome-wide genotyping data renew knowledge on genetic diversity of a worldwide alfalfa collection and give insights on genetic control of phenology traits.Front Plant Sci. 2023 Jul 5;14:1196134. doi: 10.3389/fpls.2023.1196134. eCollection 2023. Front Plant Sci. 2023. PMID: 37476178 Free PMC article.
References
-
- Tang P., Zhang Y., Sun X., Tian D., Yang S., Ding J. Disease resistance signature of the leucine-rich repeat receptor-like kinase genes in four plant species. Plant Sci. 2010;179:399–406. doi: 10.1016/j.plantsci.2010.06.017. - DOI
-
- Wang J., Liu S., Li C., Wang T., Zhang P., Chen K. PnLRR-RLK27, a novel leucine-rich repeats receptor-like protein kinase from the Antarctic moss Pohlia nutans, positively regulates salinity and oxidation-stress tolerance. PLoS ONE. 2017;12:e0172869. doi: 10.1371/journal.pone.0172869. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

