The Structural Basis of Peptide Binding at Class A G Protein-Coupled Receptors
- PMID: 35011444
- PMCID: PMC8746363
- DOI: 10.3390/molecules27010210
The Structural Basis of Peptide Binding at Class A G Protein-Coupled Receptors
Abstract
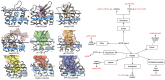
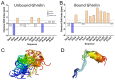
G protein-coupled receptors (GPCRs) represent the largest membrane protein family and a significant target class for therapeutics. Receptors from GPCRs' largest class, class A, influence virtually every aspect of human physiology. About 45% of the members of this family endogenously bind flexible peptides or peptides segments within larger protein ligands. While many of these peptides have been structurally characterized in their solution state, the few studies of peptides in their receptor-bound state suggest that these peptides interact with a shared set of residues and undergo significant conformational changes. For the purpose of understanding binding dynamics and the development of peptidomimetic drug compounds, further studies should investigate the peptide ligands that are complexed to their cognate receptor.
Keywords: class A GPCR; non-canonical amino acids; peptide GPCR; peptide docking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
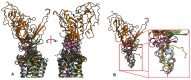

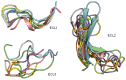





Similar articles
-
Peptide recognition, signaling and modulation of class B G protein-coupled receptors.Curr Opin Struct Biol. 2018 Aug;51:53-60. doi: 10.1016/j.sbi.2018.03.008. Epub 2018 Mar 19. Curr Opin Struct Biol. 2018. PMID: 29567494 Review.
-
7TM Domain Structure of Adhesion GPCRs.Handb Exp Pharmacol. 2016;234:43-66. doi: 10.1007/978-3-319-41523-9_3. Handb Exp Pharmacol. 2016. PMID: 27832483 Review.
-
Recognition of GPCRs by peptide ligands and membrane compartments theory: structural studies of endogenous peptide hormones in membrane environment.Biosci Rep. 2006 Apr;26(2):131-58. doi: 10.1007/s10540-006-9014-z. Biosci Rep. 2006. PMID: 16773462 Review.
-
Understanding Peptide Binding in Class A G Protein-Coupled Receptors.Mol Pharmacol. 2019 Nov;96(5):550-561. doi: 10.1124/mol.119.115915. Epub 2019 Jul 10. Mol Pharmacol. 2019. PMID: 31436539 Free PMC article. Review.
-
Structural Characterization of Receptor-Receptor Interactions in the Allosteric Modulation of G Protein-Coupled Receptor (GPCR) Dimers.Int J Mol Sci. 2021 Mar 22;22(6):3241. doi: 10.3390/ijms22063241. Int J Mol Sci. 2021. PMID: 33810175 Free PMC article. Review.
Cited by
-
Computational design of dynamic receptor-peptide signaling complexes applied to chemotaxis.Nat Commun. 2023 May 19;14(1):2875. doi: 10.1038/s41467-023-38491-9. Nat Commun. 2023. PMID: 37208363 Free PMC article.
-
A Biomimetic C-Terminal Extension Strategy for Photocaging Amidated Neuropeptides.J Am Chem Soc. 2023 Sep 13;145(36):19611-19621. doi: 10.1021/jacs.3c03913. Epub 2023 Aug 31. J Am Chem Soc. 2023. PMID: 37649440 Free PMC article.
-
Molecular docking of danuglipron uncovers potential crossovers between GLP-1R and the endocannabinoid system.MicroPubl Biol. 2025 Jul 24;2025:10.17912/micropub.biology.001690. doi: 10.17912/micropub.biology.001690. eCollection 2025. MicroPubl Biol. 2025. PMID: 40838124 Free PMC article.
-
In silico analysis of crustacean hyperglycemic hormone family G protein-coupled receptor candidates.Front Endocrinol (Lausanne). 2024 Jan 9;14:1322800. doi: 10.3389/fendo.2023.1322800. eCollection 2023. Front Endocrinol (Lausanne). 2024. PMID: 38298185 Free PMC article.
-
Orexins in apoptosis: a dual regulatory role.Front Cell Neurosci. 2024 Apr 12;18:1336145. doi: 10.3389/fncel.2024.1336145. eCollection 2024. Front Cell Neurosci. 2024. PMID: 38699177 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

