Fruit Bromelain-Derived Peptide Potentially Restrains the Attachment of SARS-CoV-2 Variants to hACE2: A Pharmacoinformatics Approach
- PMID: 35011492
- PMCID: PMC8746556
- DOI: 10.3390/molecules27010260
Fruit Bromelain-Derived Peptide Potentially Restrains the Attachment of SARS-CoV-2 Variants to hACE2: A Pharmacoinformatics Approach
Abstract
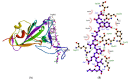
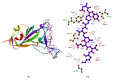
Before entering the cell, the SARS-CoV-2 spike glycoprotein receptor-binding domain (RBD) binds to the human angiotensin-converting enzyme 2 (hACE2) receptor. Hence, this RBD is a critical target for the development of antiviral agents. Recent studies have discovered that SARS-CoV-2 variants with mutations in the RBD have spread globally. The purpose of this in silico study was to determine the potential of a fruit bromelain-derived peptide. DYGAVNEVK. to inhibit the entry of various SARS-CoV-2 variants into human cells by targeting the hACE binding site within the RBD. Molecular docking analysis revealed that DYGAVNEVK interacts with several critical RBD binding residues responsible for the adhesion of the RBD to hACE2. Moreover, 100 ns MD simulations revealed stable interactions between DYGAVNEVK and RBD variants derived from the trajectory of root-mean-square deviation (RMSD), radius of gyration (Rg), and root-mean-square fluctuation (RMSF) analysis, as well as free binding energy calculations. Overall, our computational results indicate that DYGAVNEVK warrants further investigation as a candidate for preventing SARS-CoV-2 due to its interaction with the RBD of SARS-CoV-2 variants.
Keywords: COVID-19; RBD mutation; SARS-CoV-2 variants; bromelain; in silico; molecular dynamics simulation; peptide; peptide-protein interaction; receptor-binding domain.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

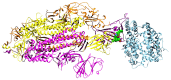
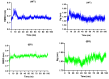

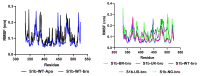
Similar articles
-
Inhibition of S-protein RBD and hACE2 Interaction for Control of SARSCoV- 2 Infection (COVID-19).Mini Rev Med Chem. 2021;21(6):689-703. doi: 10.2174/1389557520666201117111259. Mini Rev Med Chem. 2021. PMID: 33208074 Review.
-
Withanone from Withania somnifera Attenuates SARS-CoV-2 RBD and Host ACE2 Interactions to Rescue Spike Protein Induced Pathologies in Humanized Zebrafish Model.Drug Des Devel Ther. 2021 Mar 11;15:1111-1133. doi: 10.2147/DDDT.S292805. eCollection 2021. Drug Des Devel Ther. 2021. PMID: 33737804 Free PMC article.
-
De novo design, retrosynthetic analysis and combinatorial synthesis of a hybrid antiviral (VTAR-01) to inhibit the interaction of SARS-CoV2 spike glycoprotein with human angiotensin-converting enzyme 2.Biol Open. 2020 Oct 15;9(10):bio054056. doi: 10.1242/bio.054056. Biol Open. 2020. PMID: 32878881 Free PMC article.
-
Computational design of ultrashort peptide inhibitors of the receptor-binding domain of the SARS-CoV-2 S protein.Brief Bioinform. 2021 Nov 5;22(6):bbab243. doi: 10.1093/bib/bbab243. Brief Bioinform. 2021. PMID: 34180984
-
Molecular Dynamics Studies on the Structural Characteristics for the Stability Prediction of SARS-CoV-2.Int J Mol Sci. 2021 Aug 13;22(16):8714. doi: 10.3390/ijms22168714. Int J Mol Sci. 2021. PMID: 34445414 Free PMC article. Review.
Cited by
-
An integrated understanding of the evolutionary and structural features of the SARS-CoV-2 spike receptor binding domain (RBD).Int J Biol Macromol. 2022 Sep 30;217:492-505. doi: 10.1016/j.ijbiomac.2022.07.022. Epub 2022 Jul 13. Int J Biol Macromol. 2022. PMID: 35841961 Free PMC article.
-
Chemico-pharmacological and computational studies of Ophiorrhiza fasciculata D. Don and Psychotria silhetensis Hook. f. focusing cytotoxic, thrombolytic, anti-inflammatory, antioxidant, and antibacterial properties.Heliyon. 2023 Sep 13;9(9):e20100. doi: 10.1016/j.heliyon.2023.e20100. eCollection 2023 Sep. Heliyon. 2023. PMID: 37809757 Free PMC article.
-
Comparative genomics, evolutionary epidemiology, and RBD-hACE2 receptor binding pattern in B.1.1.7 (Alpha) and B.1.617.2 (Delta) related to their pandemic response in UK and India.Infect Genet Evol. 2022 Jul;101:105282. doi: 10.1016/j.meegid.2022.105282. Epub 2022 Apr 13. Infect Genet Evol. 2022. PMID: 35427787 Free PMC article.
-
Phytochemical Isolation and Antimicrobial, Thrombolytic, Anti-inflammatory, Analgesic, and Antidiarrheal Activities from the Shell of Commonly Available Citrus reticulata Blanco: Multifaceted Role of Polymethoxyflavones.Nutr Metab Insights. 2025 Apr 24;18:11786388251327668. doi: 10.1177/11786388251327668. eCollection 2025. Nutr Metab Insights. 2025. PMID: 40297736 Free PMC article.
-
Chemical and Pharmacological Profiling of Wrightia coccinea (Roxb. Ex Hornem.) Sims Focusing Antioxidant, Cytotoxic, Antidiarrheal, Hypoglycemic, and Analgesic Properties.Molecules. 2022 Jun 22;27(13):4024. doi: 10.3390/molecules27134024. Molecules. 2022. PMID: 35807270 Free PMC article.
References
-
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. - DOI - PubMed
-
- Padilla-Sanchez V. SARS-CoV-2 Structural Analysis of Receptor Binding Domain New Variants from United Kingdom and South Africa. Res. Ideas Outcomes. 2021;7:e62936. doi: 10.3897/rio.7.e62936. - DOI
-
- Albini A., Di Guardo G., Noonan D.M.C., Lombardo M. The SARS-CoV-2 receptor, ACE-2, is expressed on many different cell types: Implications for ACE-inhibitor- and angiotensin II receptor blocker-based cardiovascular therapies. Intern. Emerg. Med. 2020;15:759–766. doi: 10.1007/s11739-020-02364-6. - DOI - PMC - PubMed
-
- Lukassen S., Chua R.L., Trefzer T., Kahn N.C., Schneider M.A., Muley T., Winter H., Meister M., Veith C., Boots A.W., et al. SARS -CoV-2 receptor ACE 2 and TMPRSS 2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020;39:e105114. doi: 10.15252/embj.2020105114. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

