TNFα Mediates Inflammation-Induced Effects on PPARG Splicing in Adipose Tissue and Mesenchymal Precursor Cells
- PMID: 35011604
- PMCID: PMC8750445
- DOI: 10.3390/cells11010042
TNFα Mediates Inflammation-Induced Effects on PPARG Splicing in Adipose Tissue and Mesenchymal Precursor Cells
Abstract
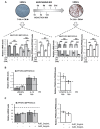
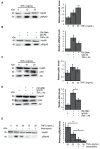
Low-grade chronic inflammation and reduced differentiation capacity are hallmarks of hypertrophic adipose tissue (AT) and key contributors of insulin resistance. We identified PPARGΔ5 as a dominant-negative splicing isoform overexpressed in the AT of obese/diabetic patients able to impair adipocyte differentiation and PPARγ activity in hypertrophic adipocytes. Herein, we investigate the impact of macrophage-secreted pro-inflammatory factors on PPARG splicing, focusing on PPARGΔ5. We report that the epididymal AT of LPS-treated mice displays increased PpargΔ5/cPparg ratio and reduced expression of Pparg-regulated genes. Interestingly, pro-inflammatory factors secreted from murine and human pro-inflammatory macrophages enhance the PPARGΔ5/cPPARG ratio in exposed adipogenic precursors. TNFα is identified herein as factor able to alter PPARG splicing-increasing PPARGΔ5/cPPARG ratio-through PI3K/Akt signaling and SRp40 splicing factor. In line with in vitro data, TNFA expression is higher in the SAT of obese (vs. lean) patients and positively correlates with PPARGΔ5 levels. In conclusion, our results indicate that inflammatory factors secreted by metabolically-activated macrophages are potent stimuli that modulate the expression and splicing of PPARG. The resulting imbalance between canonical and dominant negative isoforms may crucially contribute to impair PPARγ activity in hypertrophic AT, exacerbating the defective adipogenic capacity of precursor cells.
Keywords: PPARG splicing; TNFα; adipocyte precursors; dominant negative isoform; hypertrophic obesity; inflammation.
Conflict of interest statement
The authors declare no conflict of interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- PON Ricerca e Innovazione 2014-2020, PON Ars01_01270 "Innovative Device For SHAping the Risk of Diabetes" (IDF SHARID)/Ministero dell'Istruzione, dell'Università e della Ricerca
- Boehringer Ingelheim European Research Programme in Microvascular Complications of Diabetes/European Foundation for the Study of Diabetes
LinkOut - more resources
Full Text Sources
Research Materials

