Recognition and Chaperoning by Pex19, Followed by Trafficking and Membrane Insertion of the Peroxisome Proliferation Protein, Pex11
- PMID: 35011719
- PMCID: PMC8750153
- DOI: 10.3390/cells11010157
Recognition and Chaperoning by Pex19, Followed by Trafficking and Membrane Insertion of the Peroxisome Proliferation Protein, Pex11
Abstract
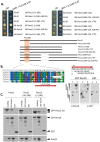
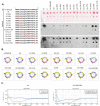
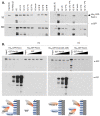
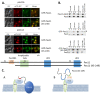
Pex11, an abundant peroxisomal membrane protein (PMP), is required for division of peroxisomes and is robustly imported to peroxisomal membranes. We present a comprehensive analysis of how the Pichia pastoris Pex11 is recognized and chaperoned by Pex19, targeted to peroxisome membranes and inserted therein. We demonstrate that Pex11 contains one Pex19-binding site (Pex19-BS) that is required for Pex11 insertion into peroxisomal membranes by Pex19, but is non-essential for peroxisomal trafficking. We provide extensive mutational analyses regarding the recognition of Pex19-BS in Pex11 by Pex19. Pex11 also has a second, Pex19-independent membrane peroxisome-targeting signal (mPTS) that is preserved among Pex11-family proteins and anchors the human HsPex11γ to the outer leaflet of the peroxisomal membrane. Thus, unlike most PMPs, Pex11 can use two mechanisms of transport to peroxisomes, where only one of them depends on its direct interaction with Pex19, but the other does not. However, Pex19 is necessary for membrane insertion of Pex11. We show that Pex11 can self-interact, using both homo- and/or heterotypic interactions involving its N-terminal helical domains. We demonstrate that Pex19 acts as a chaperone by interacting with the Pex19-BS in Pex11, thereby protecting Pex11 from spontaneous oligomerization that would otherwise cause its aggregation and subsequent degradation.
Keywords: Pex11; Pex19; peroxisomal membrane protein; peroxisome division; peroxisome proliferation protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures