Targeting PGM3 as a Novel Therapeutic Strategy in KRAS/LKB1 Co-Mutant Lung Cancer
- PMID: 35011738
- PMCID: PMC8750012
- DOI: 10.3390/cells11010176
Targeting PGM3 as a Novel Therapeutic Strategy in KRAS/LKB1 Co-Mutant Lung Cancer
Abstract
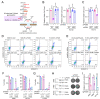
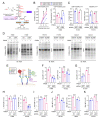
In non-small-cell lung cancer (NSCLC), concurrent mutations in the oncogene KRAS and tumor suppressor STK11 (also known as LKB1) confer an aggressive malignant phenotype, an unfavourability towards immunotherapy, and overall poor prognoses in patients. In a previous study, we showed that murine KRAS/LKB1 co-mutant tumors and human co-mutant cancer cells have an enhanced dependence on glutamine-fructose-6-phosphate transaminase 2 (GFPT2), a rate-limiting enzyme in the hexosamine biosynthesis pathway (HBP), which could be targeted to reduce survival of KRAS/LKB1 co-mutants. Here, we found that KRAS/LKB1 co-mutant cells also exhibit an increased dependence on N-acetylglucosamine-phosphate mutase 3 (PGM3), an enzyme downstream of GFPT2. Genetic or pharmacologic suppression of PGM3 reduced KRAS/LKB1 co-mutant tumor growth in both in vitro and in vivo settings. Our results define an additional metabolic vulnerability in KRAS/LKB1 co-mutant tumors to the HBP and provide a rationale for targeting PGM3 in this aggressive subtype of NSCLC.
Keywords: KRAS and LKB1 co-mutations; O-GlcNAcylation; cancer metabolism; glycosylation; metabolic vulnerability; non-small cell lung cancer; the hexosamine biosynthesis pathway.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
The hexosamine biosynthesis pathway is a targetable liability in KRAS/LKB1 mutant lung cancer.Nat Metab. 2020 Dec;2(12):1401-1412. doi: 10.1038/s42255-020-00316-0. Epub 2020 Nov 30. Nat Metab. 2020. PMID: 33257855 Free PMC article.
-
Downregulation of GFPT2 enhances cisplatin chemotherapy sensitivity in STK11/KRAS mutant non-small cell lung cancer by regulating the hexosamine biosynthesis pathway, resisting tumor growth.Cytokine. 2025 Jul;191:156943. doi: 10.1016/j.cyto.2025.156943. Epub 2025 Apr 19. Cytokine. 2025. PMID: 40253947
-
Co-occurring KRAS mutation/LKB1 loss in non-small cell lung cancer cells results in enhanced metabolic activity susceptible to caloric restriction: an in vitro integrated multilevel approach.J Exp Clin Cancer Res. 2018 Dec 4;37(1):302. doi: 10.1186/s13046-018-0954-5. J Exp Clin Cancer Res. 2018. PMID: 30514331 Free PMC article.
-
KRAS-Mutant non-small cell lung cancer: From biology to therapy.Lung Cancer. 2018 Oct;124:53-64. doi: 10.1016/j.lungcan.2018.07.013. Epub 2018 Jul 19. Lung Cancer. 2018. PMID: 30268480 Review.
-
Efficacy of immunotherapy in KRAS-mutant non-small-cell lung cancer with comutations.Immunotherapy. 2021 Aug;13(11):941-952. doi: 10.2217/imt-2021-0090. Epub 2021 Jun 11. Immunotherapy. 2021. PMID: 34114474 Review.
Cited by
-
Daily Practice Assessment of KRAS Status in NSCLC Patients: A New Challenge for the Thoracic Pathologist Is Right around the Corner.Cancers (Basel). 2022 Mar 23;14(7):1628. doi: 10.3390/cancers14071628. Cancers (Basel). 2022. PMID: 35406400 Free PMC article. Review.
-
Identification of hexosamine biosynthesis pathway as a novel prognostic signature and its correlation with immune infiltration in bladder cancer.Front Mol Biosci. 2022 Sep 8;9:1009168. doi: 10.3389/fmolb.2022.1009168. eCollection 2022. Front Mol Biosci. 2022. PMID: 36158580 Free PMC article.
-
The Hexosamine Biosynthesis Pathway: Regulation and Function.Genes (Basel). 2023 Apr 18;14(4):933. doi: 10.3390/genes14040933. Genes (Basel). 2023. PMID: 37107691 Free PMC article. Review.
-
O-linked β-N-acetylglucosaminylation in lung cancer and beyond: A multidimensional perspective.Chin Med J (Engl). 2025 Feb 5;138(3):355-357. doi: 10.1097/CM9.0000000000003329. Epub 2024 Dec 31. Chin Med J (Engl). 2025. PMID: 39787372 Free PMC article. No abstract available.
-
Meta-analysis of the effect of PGM on survival prognosis of tumor patients.Front Oncol. 2022 Dec 5;12:1060372. doi: 10.3389/fonc.2022.1060372. eCollection 2022. Front Oncol. 2022. PMID: 36544711 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

